Team:Cambridge/Experiments/Initial Exercise Group A
From 2011.igem.org
(→Restriction Mapping) |
|||
| (9 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
==Initial Exercise: Cat, Jonathan, Haydn and Ai== | ==Initial Exercise: Cat, Jonathan, Haydn and Ai== | ||
| - | As a 'warm-up' exercise to acquaint the group with molecular biological laboratory techniques, three mini-teams were tasked with creating an interesting GFP fusion. Group A decided that visualing ftsZ in real time, in vivo would be | + | As a 'warm-up' exercise to acquaint the group with molecular biological laboratory techniques, three mini-teams were tasked with creating an interesting GFP fusion. Group A decided that visualing ftsZ in real time, in vivo would be good fun. |
| - | Ftsz was first identified in a mutant screen in 1980 | + | Ftsz was first identified in a mutant screen in 1980 (Lutkenhaus, Wolf-Watz and Donachie) as a gene recquired for bacterial cytokinesis (cell division). It is known that a ring structure partly composed of this protein forms around the cell equator at division. It was hoped that a glowing green ring would therefore be visualisable through confocal microscopy as a result of our fusion. |
==Notes== | ==Notes== | ||
| Line 21: | Line 21: | ||
===Returned Primers=== | ===Returned Primers=== | ||
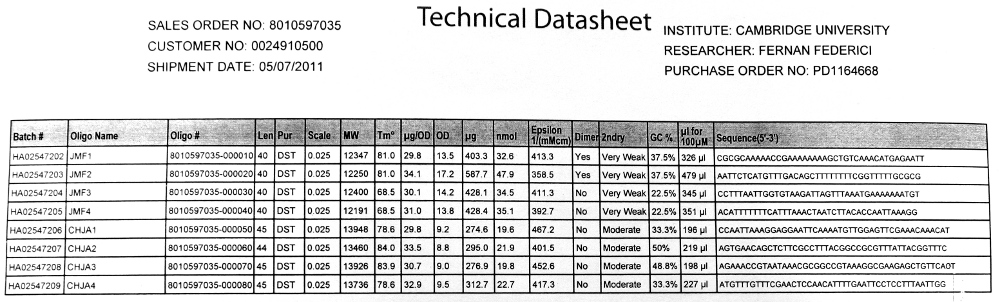
| - | [[File:CAM HJCA PRIMERS.JPG | thumb | | + | [[File:CAM HJCA PRIMERS.JPG | thumb | 720px | center | The Technical datasheet we received]] |
==Procedures== | ==Procedures== | ||
| Line 40: | Line 40: | ||
| - | ===2. GFP side=== | + | ====2. GFP side==== |
Amplify fragment containing GFP coding sequence (RHS on diagram above) & part of vector plasmid | Amplify fragment containing GFP coding sequence (RHS on diagram above) & part of vector plasmid | ||
| Line 52: | Line 52: | ||
| - | ===3. Promoter side=== | + | ====3. Promoter side==== |
Amplify fragment containing promoter (LHS on above) | Amplify fragment containing promoter (LHS on above) | ||
| Line 63: | Line 63: | ||
|} | |} | ||
| + | ==Restriction Mapping== | ||
| + | The nucleotide sequence of our insert and the plasmid used were obtained from the literature. These were collated in the plasmid-editing program 'aPe' and all restriction sites for which enzymes were available mapped. | ||
| + | [[File:team1.jpg|thumb]] | ||
| + | A particular enzyme was chosen from the map that conveniently cut in three places to give fragments of a good size to be resolved on a gel. The expected gel results were modelled using the same program. | ||
| + | Our Gibson assembly unfortunately failed, and with the time constraint of the Jamboree looming even at this early stage, the initial project had to be abandoned. | ||
| + | [[File:cam_team1a.jpg|thumb]] | ||
{{Template:Team:Cambridge/CAM_2011_TEMPLATE_FOOT}} | {{Template:Team:Cambridge/CAM_2011_TEMPLATE_FOOT}} | ||
Latest revision as of 13:43, 21 September 2011
Contents |
Initial Exercise: Cat, Jonathan, Haydn and Ai
As a 'warm-up' exercise to acquaint the group with molecular biological laboratory techniques, three mini-teams were tasked with creating an interesting GFP fusion. Group A decided that visualing ftsZ in real time, in vivo would be good fun.
Ftsz was first identified in a mutant screen in 1980 (Lutkenhaus, Wolf-Watz and Donachie) as a gene recquired for bacterial cytokinesis (cell division). It is known that a ring structure partly composed of this protein forms around the cell equator at division. It was hoped that a glowing green ring would therefore be visualisable through confocal microscopy as a result of our fusion.
Notes
Primer Design
The plasmid vector we were supplied with contains a strong promoter upstream of a sfGFP coding sequence. Our fusion design relies on amplifying the ftsZ coding region from Bacillus and creating regions of overlap between this and the GFP coding sequence in the plasmid, in order to create the gene fusion.
The desired end product is shown below, with the plasmid in lowercase and the ftsZ coding region insert in upper case.
ccaattaaaggaggaattcaaaATGTTGGAGTTCGAAACAAACAT-----AGAAACCGTAATAAACGCGGCcgtaaaggcgaagagctgttcact ggttaatttcctccttaagtttTACAACCTCAAGCTTTGTTTGTA-----TCTTTGGCATTATTTGCGCCGgcatttccgcttctcgacaagtga
Returned Primers
Procedures
PCR
Three separate PCR reaction were required, detailed below. These reactions were carried out using the automatic PCR machine.
1. ftsZ
Aim is to amplify ftsZ.
| Template | B. subtilus genome |
| FWD Primer | ccaattaaaggaggaattcaaaATGTTGGAGTTCGAAACAAACAT Order Code: 050 |
| REV Primer | agtgaacagctcttcgcctttacgGCCGCGTTTATTACGGTTTC Order Code: 060 |
2. GFP side
Amplify fragment containing GFP coding sequence (RHS on diagram above) & part of vector plasmid
| Template | Vector Plasmid. |
| FWD Primer | AGAAACCGTAATAAACGCGGCcgtaaaggcgaagagctgttcact Order Code: 070 |
| REV Primer | provided Code: B |
3. Promoter side
Amplify fragment containing promoter (LHS on above)
| Template | Vector Plasmid. |
| FWD Primer | provided Code: A |
| REV Primer | ATGTTTGTTTCGAACTCCAACATtttgaattcctcctttaattgg Order Code: 080 |
Restriction Mapping
The nucleotide sequence of our insert and the plasmid used were obtained from the literature. These were collated in the plasmid-editing program 'aPe' and all restriction sites for which enzymes were available mapped.
A particular enzyme was chosen from the map that conveniently cut in three places to give fragments of a good size to be resolved on a gel. The expected gel results were modelled using the same program.
Our Gibson assembly unfortunately failed, and with the time constraint of the Jamboree looming even at this early stage, the initial project had to be abandoned.
 "
"