Team:Paris Bettencourt/Experiment/T7 diff subt subt microfluidic
From 2011.igem.org
| (13 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
<html> | <html> | ||
| - | <h1> | + | <h1>Nanotube-assisted diffusion of T7 polymerase in Microfluidics</h1> |
| + | <h2>Experimental Scheme</h2> | ||
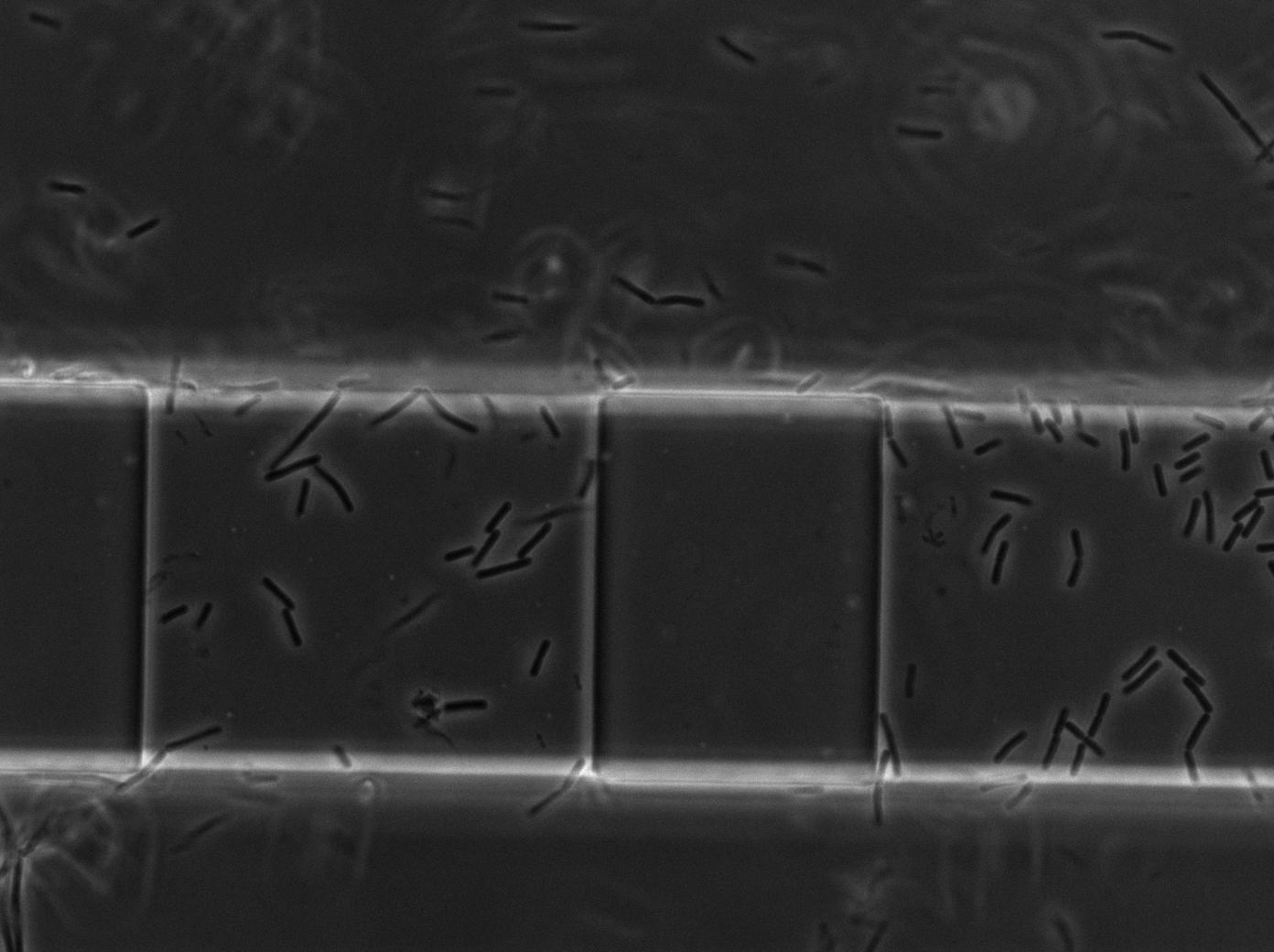
| + | <p>In order to test whether our <i>Bacillus subtilis</i> T7 emitters & receivers can form nanotubes when mixed, we mix them in microfluidic system modified from Jeff Hasty's recent <a href="http://biodynamics.ucsd.edu/pubs/articles/Mondragon11.pdf">paper</a> <a href="https://2011.igem.org/Team:Paris_Bettencourt/Experiments/Methodologies/Microchemostat_HastyJ#references">[1]</a>, shown below. For more detailed information regarding the microfluidic device and experimental procedure, check our <a href="https://2011.igem.org/Team:Paris_Bettencourt/Experiments/Methodologies/Microchemostat_HastyJ">methodology page</a>. </p> | ||
| + | |||
| + | <p><center><a href="https://2011.igem.org/File:Paris_microchemostat_channels_and_chambers.jpg"><img height=540px align="center" src="https://static.igem.org/mediawiki/2011/f/f9/Paris_microchemostat_channels_and_chambers.jpg"></a></center></p> | ||
| + | |||
| + | <p>We imaged two channels: one experimental channel injected with an emitter strain (RFP constitutive) and a receiver strain (pT7-T7polyermase-GFP); the other is a control channel injected with only the receiver strain. The receiver strain contains the T7 autoloop, which will gain a strong fluorescence when activated. </p> | ||
| + | |||
| + | <h2>Results</h2> | ||
| + | |||
| + | <p>The receiver strain by itself in our control channel have never produced any GFP positive cells, in contrast to the leaky situation where the T7 autoloop is encoded by a plasmid in E.coli. This is due to the low copy number of T7 polymerase gene on the chromosome. Yet the channel with the mixed population start to produce GFP positive cells immediately after injection. Yet due to the large population in the main channel compared to our single layer chambers, we never actually saw a turning-on event. But we do see a large increase of GFP+ cells in the mixed channel, where there are 0 GFP+ cells in the control channel.</p> | ||
| + | |||
| + | </html><center> | ||
| + | {| border="1" class="wikitable" style="text-align: center;" | ||
| + | |+Example of the GFP+ cells in the mix channel at around 3h after injection | ||
| + | |- | ||
| + | |[[File:Microchemostat_BST7_emit-RFP_autoloop-GFP3_w1TRANS_s5_t62.jpg|450px|thumb|center| Position5 186min trans image]] | ||
| + | |[[File:Microchemostat_BST7_emit-RFP_autoloop-GFP3_w2GFP_s5_t62.jpg|450px|thumb|center| Position5 186min gfp image]] | ||
| + | |} | ||
| + | |||
| + | <i>GFP+ cells washed down from the main flow channel</i></center> | ||
| + | |||
| + | <html> | ||
| + | |||
| + | <p>We think what's happening here is that there are biofilm-like populations stuck at the entrance of the channels; And receiver cells turn on the T7 autoloop in this population in the mix channel, and the increase of GFP+ cells are descents of those cells that are originally turned on washed downstream by the flow. Since the cells that are washed downstream is not in monolayer thus not very well focused, we decided to quantify this by integrating the GFP signals in the whole image. Below is the summary of the total GFP fluorescent signals in 3 imaging positions in each channel. For each GFP image at each time point in each position, we cut off the image at a rather stringent threshold, so that the background is decreased to zero, and then we integrated the GFP signal left-over in the image to represent the amount of GFP+ receiver in that image.</p> | ||
| + | |||
| + | <p><center><a href="https://static.igem.org/mediawiki/2011/6/69/Paris2011_T7-microfluidics-gfpsums.jpg"><img height=540px align="center" src="https://static.igem.org/mediawiki/2011/6/69/Paris2011_T7-microfluidics-gfpsums.jpg"></a></center></p> | ||
| + | |||
| + | <h2>Conclusion</h2> | ||
| + | |||
| + | <p>The fact that when mixed with emitters, the receiver cells can turn on GFP signals in such large numbers and in such short time, indicates:</p> | ||
| + | |||
| + | <p><br>1. Our pT7-T7polyermase-GFP construct worked when integrated into <i>bacillus subtilis</i> chromosome.</br> | ||
| + | <br>2. We can't find another explanation other than nanotube-assisted diffusion of T7 polymerase in the biofilm in the main channel for the turning on events we saw in the channels. | ||
| + | </p> | ||
| + | |||
| + | <div id="citation_box"> | ||
| + | <p id="references">References</p> | ||
| + | <ol> | ||
| + | |||
| + | <li><i> Entrainment of a population of synthetic genetic oscillators.</i> Mondragón-Palomino, O., Danino, T., Selimkhanov, J., Tsimring, L. & Hasty, J. Science 333, 1315-1319 (2011).</li> | ||
| + | |||
| + | </ol> | ||
| + | </div> | ||
| + | <br> | ||
<!-- PAGE FOOTER -- ITEMS FROM COLUMN ! HAVE BEEN MOVED HERE -- RDR --> | <!-- PAGE FOOTER -- ITEMS FROM COLUMN ! HAVE BEEN MOVED HERE -- RDR --> | ||
Latest revision as of 03:53, 29 October 2011

Nanotube-assisted diffusion of T7 polymerase in Microfluidics
Experimental Scheme
In order to test whether our Bacillus subtilis T7 emitters & receivers can form nanotubes when mixed, we mix them in microfluidic system modified from Jeff Hasty's recent paper [1], shown below. For more detailed information regarding the microfluidic device and experimental procedure, check our methodology page.

We imaged two channels: one experimental channel injected with an emitter strain (RFP constitutive) and a receiver strain (pT7-T7polyermase-GFP); the other is a control channel injected with only the receiver strain. The receiver strain contains the T7 autoloop, which will gain a strong fluorescence when activated.
Results
The receiver strain by itself in our control channel have never produced any GFP positive cells, in contrast to the leaky situation where the T7 autoloop is encoded by a plasmid in E.coli. This is due to the low copy number of T7 polymerase gene on the chromosome. Yet the channel with the mixed population start to produce GFP positive cells immediately after injection. Yet due to the large population in the main channel compared to our single layer chambers, we never actually saw a turning-on event. But we do see a large increase of GFP+ cells in the mixed channel, where there are 0 GFP+ cells in the control channel.
We think what's happening here is that there are biofilm-like populations stuck at the entrance of the channels; And receiver cells turn on the T7 autoloop in this population in the mix channel, and the increase of GFP+ cells are descents of those cells that are originally turned on washed downstream by the flow. Since the cells that are washed downstream is not in monolayer thus not very well focused, we decided to quantify this by integrating the GFP signals in the whole image. Below is the summary of the total GFP fluorescent signals in 3 imaging positions in each channel. For each GFP image at each time point in each position, we cut off the image at a rather stringent threshold, so that the background is decreased to zero, and then we integrated the GFP signal left-over in the image to represent the amount of GFP+ receiver in that image.

Conclusion
The fact that when mixed with emitters, the receiver cells can turn on GFP signals in such large numbers and in such short time, indicates:
1. Our pT7-T7polyermase-GFP construct worked when integrated into bacillus subtilis chromosome.
2. We can't find another explanation other than nanotube-assisted diffusion of T7 polymerase in the biofilm in the main channel for the turning on events we saw in the channels.
References
- Entrainment of a population of synthetic genetic oscillators. Mondragón-Palomino, O., Danino, T., Selimkhanov, J., Tsimring, L. & Hasty, J. Science 333, 1315-1319 (2011).
 "
"