Team:Potsdam Bioware/Project/Details Selection
From 2011.igem.org
(→Construction of two component for the in vivo selection assay) |
(→Introduction) |
||
| (72 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
==<i>In Vivo</i> Selection== | ==<i>In Vivo</i> Selection== | ||
===Introduction=== | ===Introduction=== | ||
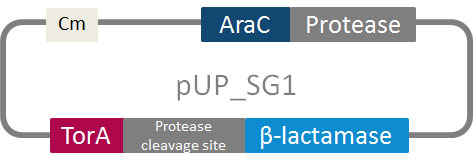
| - | In addition to the phage display we developed a novel selection system. The design aimed for a | + | In addition to the phage display we developed a novel selection system. The design aimed for a inexpensive and time-saving alternative in contrast to an <i>in vitro</i> screen of protease inhibition. The assay allows us to select effective inhibitors for any protease, among the billions of randomly generated mutants of the Microviridin. For this purpose we designed a plasmid containing two devices, first a protease activity detector and second a protease generator. For the protease activity detector we modified the TAT-hitchhiker system developed by DeLisa <i>et al.</i> |
| - | We used the BioBrick <partinfo>I757010</partinfo> (β-lactamase) as well as <partinfo>K208005</partinfo> (ssTorA) and fused them together via a linker peptide. For the protease generator we cloned the | + | We used the BioBrick <partinfo>I757010</partinfo> (β-lactamase) as well as <partinfo>K208005</partinfo> (ssTorA) and fused them together via a linker peptide. For the protease generator we cloned the arabinose induction system in front of a protease in iGEM standard.<br> |
[[File:UP_images_TorA-construct-design2_28.10.2011.png.jpg|center|500px|thumb|'''Figure 1:''' Schema of our constructed selection vector containing protease generator and protease activity detector]]<br> | [[File:UP_images_TorA-construct-design2_28.10.2011.png.jpg|center|500px|thumb|'''Figure 1:''' Schema of our constructed selection vector containing protease generator and protease activity detector]]<br> | ||
| - | The system works in a combined manner of the two devices. In order to confer β-lactam antibiotic resistance to cells the β-lactamase has to be exported in the periplasm. This transport is mediated by the TorA export sequence via the Twin-Arginine Translocation (TAT) system (DeLisa, | + | The system works in a combined manner of the two devices. In order to confer β-lactam antibiotic resistance to cells the β-lactamase has to be exported in the periplasm. This transport is mediated by the TorA export sequence via the Twin-Arginine Translocation (TAT) system (DeLisa, 2009). The linker peptide between the TorA export sequence and the β-lactamase displays the corresponding protease cleavage site. In addition the linker peptide is chosen as short as possible to imitate folding because the TAT pathway only allows transport of correctly folded substrates. The protease as well as the linker peptide are designed as exchangeable parts.<br> |
| - | If the protease device is functional, it will cleave the linker peptide between TorA and the β-lactamase construct which leaves the cell without any antibiotic resistance. The construct was tested with increasing | + | If the protease device is functional, it will cleave the linker peptide between TorA and the β-lactamase construct which leaves the cell without any antibiotic resistance. The construct was tested with increasing ampicillin concentrations. The number of surviving colonies was depended on the export rate of the β-lactamase into the periplasm. A high cleavage rate of the linker peptide leads to a reduced ampicillin resistance. With expressed protease a dramatically drop of the number of surviving colonies could be observed.<br> |
| - | The survival assay was carried out with and without expressed protease. By increasing of the | + | The survival assay was carried out with and without expressed protease. By increasing of the ampicillin concentration we could detect a cutoff ampicillin concentration. The colony forming units (CFU) were counted and compared. To detected whether our constructed library is able to block the protease we co-transformed the library into <i>E. coli</i> carrying our pUP_SG1 plasmid.<br> |
| - | ===Site-directed mutagenesis of | + | ===Site-directed mutagenesis of Tobacco Etch Virus and 14_3C proteases in iGEM standard=== |
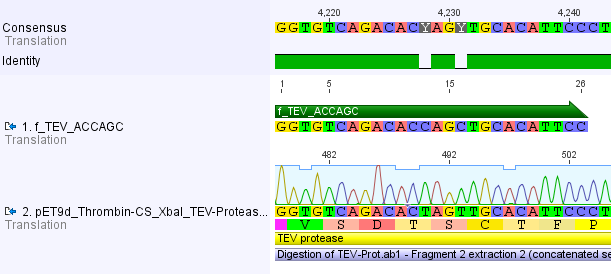
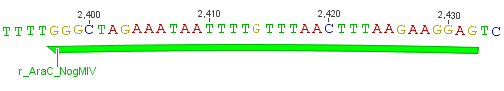
We chose two model proteases, the Tobacco Etch Virus protease, further along called TEV, and the 14_3C protease from the human rhinovirus, also known as PreScission. Both proteases contained iGEM restriction sites, one in case of the TEV and three for 14 3C, respectively. <br> | We chose two model proteases, the Tobacco Etch Virus protease, further along called TEV, and the 14_3C protease from the human rhinovirus, also known as PreScission. Both proteases contained iGEM restriction sites, one in case of the TEV and three for 14 3C, respectively. <br> | ||
| - | [[File:UP_Pic_SG_Pre-Complete.png|center|800px|thumb|'''Figure 2:''' | + | [[File:UP_Pic_SG_Pre-Complete.png|center|800px|thumb|'''Figure 2:''' Complete sequence alignment of the 14_3C protease]] |
| - | [[File:UP_Pic_SG_TEV-Complete.png|center|800px|thumb|'''Figure 3:''' | + | [[File:UP_Pic_SG_TEV-Complete.png|center|800px|thumb|'''Figure 3:''' Complete sequence alignment of the 14_3C protease]] |
<br><br> | <br><br> | ||
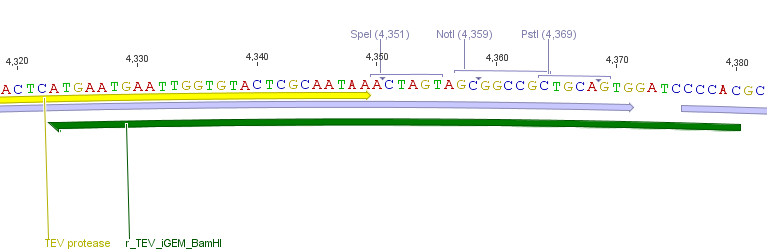
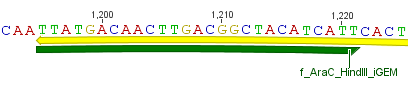
Thus, site-directed mutagenesis was applied to mutate each restriction site in E. coli. For every mutation a forward and a reverse primer had to be designed. The following pictures show the forward primers for the side directed mutagenesis, the reverse primers are the reverse complement sequence of the forward primers.<br> | Thus, site-directed mutagenesis was applied to mutate each restriction site in E. coli. For every mutation a forward and a reverse primer had to be designed. The following pictures show the forward primers for the side directed mutagenesis, the reverse primers are the reverse complement sequence of the forward primers.<br> | ||
For the 14_3C protease: | For the 14_3C protease: | ||
| - | [[File:UP_Pic_SG_Pre-End.png|left|400px|thumb|'''Figure 4:'''Reverse primer to add the iGEM suffix on the HRV 14_3C protease]] | + | [[File:UP_Pic_SG_Pre-End.png|left|400px|thumb|'''Figure 4:''' Reverse primer to add the iGEM suffix on the HRV 14_3C protease]] |
| - | [[File:UP_Pic_SG_PreMutaSpeI.png|right|400px|thumb|'''Figure 5:'''Forward primer to mutate the SpeI restriction site in the gene sequence of the HRV 14_3C protease]] | + | [[File:UP_Pic_SG_PreMutaSpeI.png|right|400px|thumb|'''Figure 5:''' Forward primer to mutate the SpeI restriction site in the gene sequence of the HRV 14_3C protease]] |
| - | [[File:UP_Pic_SG_PreMutaXbaI-1.png|left|400px|thumb|'''Figure 6:'''Forward primer to mutate the first XbaI restriction site in the gene sequence of the HRV 14_3C protease]] | + | [[File:UP_Pic_SG_PreMutaXbaI-1.png|left|400px|thumb|'''Figure 6:''' Forward primer to mutate the first XbaI restriction site in the gene sequence of the HRV 14_3C protease]] |
| - | [[File:UP_Pic_SG_PreMutaXbaI-2.png|right|400px|thumb|'''Figure 7:'''Forward primer to mutate the second XbaI restriction site in the gene sequence of the HRV 14_3C protease]] | + | [[File:UP_Pic_SG_PreMutaXbaI-2.png|right|400px|thumb|'''Figure 7:''' Forward primer to mutate the second XbaI restriction site in the gene sequence of the HRV 14_3C protease]] |
[[File:UP_Pic_SG_Pre-Start.png|left|400px|thumb|'''Figure 8:''' Forward primer to add the iGEM prefix to the HRV 14_3C protease]] | [[File:UP_Pic_SG_Pre-Start.png|left|400px|thumb|'''Figure 8:''' Forward primer to add the iGEM prefix to the HRV 14_3C protease]] | ||
| Line 33: | Line 33: | ||
[[File:UP_Pic_SG_TEV-End.png|left|400px|thumb|'''Figure 9:''' Reverse primer to add the iGEM suffix to the TEV protease]] | [[File:UP_Pic_SG_TEV-End.png|left|400px|thumb|'''Figure 9:''' Reverse primer to add the iGEM suffix to the TEV protease]] | ||
[[File:UP_Pic_SG_TEVMutaSpeI.png|right|400px|thumb|'''Figure 10:''' Forward primer to mutate the SpeI restriction site in the gene sequence of the TEV protease]] | [[File:UP_Pic_SG_TEVMutaSpeI.png|right|400px|thumb|'''Figure 10:''' Forward primer to mutate the SpeI restriction site in the gene sequence of the TEV protease]] | ||
| - | [[File:UP_Pic_SG_TEV-Start.png|left|400px|thumb|'''Figure 11:'''Forward primer to add the iGEM prefix to the TEV protease]] | + | [[File:UP_Pic_SG_TEV-Start.png|left|400px|thumb|'''Figure 11:''' Forward primer to add the iGEM prefix to the TEV protease]] |
<br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | <br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br><br> | ||
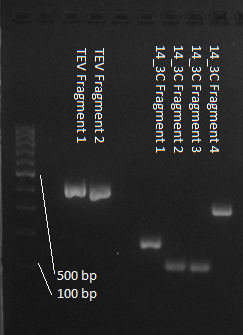
| - | We used assembly PCR to gain the final product. In case of | + | We used assembly PCR to gain the final product. In case of 14_3C two assembly PCRs had to be done. The expected size of the fragments are shown in the gel picture. In the following picture all fragments contain the correct sizes. |
<br> | <br> | ||
| - | [[File:UP_Fragments_14_3C_TEV.png|center|200px|thumb|'''Figure 12:'''Picture of the, via PCR amplified and mutated Fragment of both protease]] | + | [[File:UP_Fragments_14_3C_TEV.png|center|200px|thumb|'''Figure 12:''' Picture of the, via PCR amplified and mutated Fragment of both protease]] |
<br> | <br> | ||
===Construction of two component for the ''in vivo'' selection assay=== | ===Construction of two component for the ''in vivo'' selection assay=== | ||
| - | The ''in vivo'' assay is based on two devices. On the one hand the protease activity detector device, on the other a protease generator device. The detector device is based on the enzyme β-lactamase, which is only active inside the periplasm and provides the resistance against lactam antibiotics like | + | The ''in vivo'' assay is based on two devices. On the one hand the protease activity detector device, on the other a protease generator device. The detector device is based on the enzyme β-lactamase, which is only active inside the periplasm and provides the resistance against lactam antibiotics like ampicillin and penicillin. For the protease generator device we needed a second induction system on our plasmid, which is quite uncommon for any plasmid. So several problems had to solved to set up an effective ''in vivo'' selection assay: |
| - | *a second, very | + | *a second, very tight induction system has to be found |
| - | *a way to control the export rate of β-lactamase into the periplasm, which can be | + | *a way to control the export rate of β-lactamase into the periplasm, which can be reduced by the protease |
| - | + | We solved these problems by putting the protease detector device under the control of the lac-operon. The protease device is induced by the arabinose system of the pBAD_iGEMexpress vector. The idea to control the survival of the cells via the amount of exported protein is inspired by the Hitchhiker assay of DeLisa ''et al''. Using the TAT pathway for the export of β-lactamase into the periplasm allows us to introduce a protease specific cleavage site in front of the signal sequence regulating the export. | |
====Introducing the specific protease cleavage site==== | ====Introducing the specific protease cleavage site==== | ||
| + | To adapt the protease activity detector device to any protease we want to screen, we had to modularize it. Our detector device was set up on a vector for the Hitchhiker selection. The plasmid offers two unique restriction sites on the plasmid, after the signal sequence of TorA one restriction site for <i>Xho</i>I and a <i>Nhe</i>I restriction site in front of the β-lactamase. Both were used to introduce the specific protease cleavage site in between the TorA signal sequence and the β-lactamase. By designing two complementary oligonucleotides containing the specific protease cleavage site flanked by two short linker regions. After the digest of the vector with <i>Xho</i>I and <i>Nhe</i>I the hybridized oligonucleotides, containing the overhangs for <i>Xho</i>I and <i>Nhe</i>I can be ligated into the vector. | ||
====Adding the arabinose inducible induction system to control the proteases, TEV and 14_3C==== | ====Adding the arabinose inducible induction system to control the proteases, TEV and 14_3C==== | ||
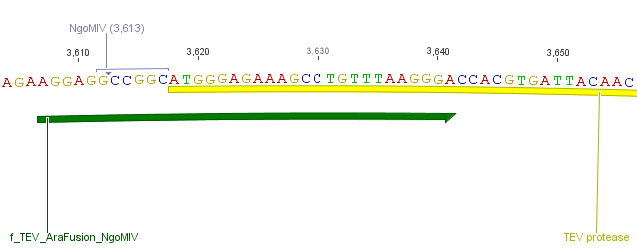
| - | We needed a time independent induction of the | + | We needed a time independent induction of the protease device. Therefore we cloned the arabionse induction system (<i>araC</i>) in front of the protease device out of the pBAD_iGEMexpress vector. The fusion of gene construct and induction system was always performed via an <i>NgoM</i>IV restriction site. This restriction site was introduced with our amplification primers. |
[[File:UP_Pic_SG_AraC-End.png|center|800px|thumb|'''Figure 13:''' Reverse primer for the amplification of the arabinose inducible induction system]] | [[File:UP_Pic_SG_AraC-End.png|center|800px|thumb|'''Figure 13:''' Reverse primer for the amplification of the arabinose inducible induction system]] | ||
[[File:UP_Pic_SG_AraC-Start.png|center|1400px|thumb|'''Figure 14:''' Forward primer for the amplification of the arabinose inducible induction system]] | [[File:UP_Pic_SG_AraC-Start.png|center|1400px|thumb|'''Figure 14:''' Forward primer for the amplification of the arabinose inducible induction system]] | ||
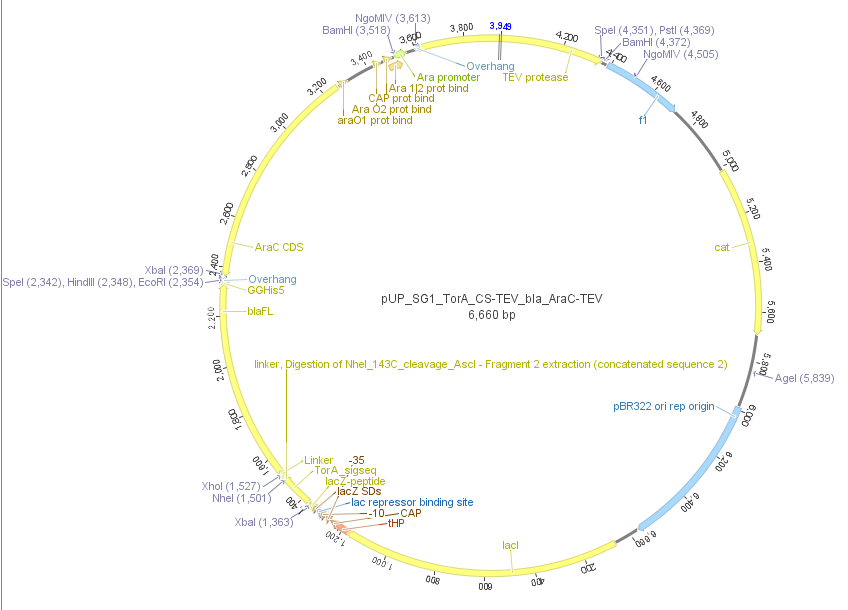
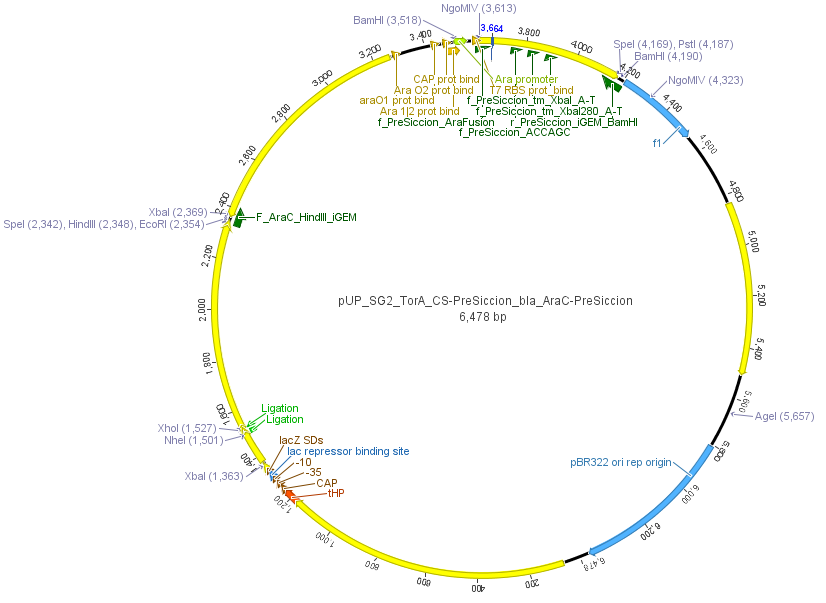
| - | We used the RFC 23 cloning strategy to get a fusion protein of our desired protease and the induction system, which allows us to induce the protease. Both genes were amplified with PCR using two special primers generating a | + | We used the RFC 23 cloning strategy to get a fusion protein of our desired protease and the induction system, which allows us to induce the protease. Both genes were amplified with PCR using two special primers generating a <i>NgoM</i>IV restrictioin site at the end of the induction system and in front of the protease. The amplified protease were digested with <i>BamH</i>I and <i>NgoM</i>IV, the arabinose inducible induction system with <i>NgoM</i>IV and <i>Hind</i>III. The vector, which contains the ssTorA_CS-Protease_β-lactamase device was digested with <i>Hind</i>III and <i>BamH</i>I. A triple ligation yielded the final construct with both devices: the protease detector and the protease generator. The two pictures below show the final vector constructs , which where used for the survival screening. |
[[File:UP_Pic_SG_TEV_SG1-Plasmid.png|center|800px|thumb|'''Figure 15:''' Final vector construct containing arabinose inducible TEV protease fusion and the protease detector device]] | [[File:UP_Pic_SG_TEV_SG1-Plasmid.png|center|800px|thumb|'''Figure 15:''' Final vector construct containing arabinose inducible TEV protease fusion and the protease detector device]] | ||
[[File:UP_Pic_SG_PreSciccion_SG2-Plasmid.png|center|800px|thumb|'''Figure 16:''' Final vector construct containing arabinose inducible HRV 14_3C protease fusion and the protease detector device]] | [[File:UP_Pic_SG_PreSciccion_SG2-Plasmid.png|center|800px|thumb|'''Figure 16:''' Final vector construct containing arabinose inducible HRV 14_3C protease fusion and the protease detector device]] | ||
| Line 63: | Line 64: | ||
<p> | <p> | ||
| - | Two major points have to be demonstrated: First, the export of β-lactamase over the TAT pathway has to be efficient enough to provide a certain resistance against | + | Two major points have to be demonstrated: First, the export of β-lactamase over the TAT pathway has to be efficient enough to provide a certain resistance against ampicillin and second, the activated protease is able to intimately reduce the resistance against ampicillin by cleavage of the TorA signal sequence from the β-lactamase.</p> |
====Testing the export efficiency of the protease activity detector==== | ====Testing the export efficiency of the protease activity detector==== | ||
| - | To test the export of the β-lactamse | + | To test the export rate of the β-lactamse of the protease activity detector device we grew an <i>E. coli</i> culture up to an OD<sub>600</sub> of 0.6. The cells were then diluted to an OD of 0.002 and 100 µL were plated in triplicates with increasing ampicillin concentrations. The graph below shows the amount of surviving cells in percentage. <br> |
| - | + | [[Image:UP_Survial-screen_bla-construct.png|center|600px|thumb|'''Figure 17:''' Amount of survived cells. The construct was induced with 1 mM IPTG. The rate of surviving cells without IPTG and without ampicilin was set to 100%. ]] | |
| - | [[Image:UP_Survial-screen_bla-construct.png|center|600px|thumb|'''Figure 17:''' Amount of survived cells. The construct was induced with IPTG. The rate of surviving cells without IPTG was set to 100%. ]] | + | |
| - | + | [[Image:UP_TEV-Survival.png|center|600px|thumb|'''Figure 18:''' Survival test at different ampicillin concentrations: A) 1000 cfu plated on agar plates containing chloramphenicol; B) Reduced amount of <i>E.coli</i> colonies, 1000 cfu were plated on agar plates containing chloramphenicol, 1mM IPTG to induce ampicillin resistance via BBa_K627012 and 400µg/mL ampicillin; C) no culture growth of <i>E.coli</i> colonies, 1000 cfu were plated on agar plates containing chloramphenicol, 1mM IPTG to induce ampicillin resistance via BBa_K627012 and 600µg/mL ampicillin]] | |
| + | [[Image:UP_14_3C_SurvTest.png|center|600px|thumb|'''Figure 19:''' Survival test at different ampicillin concentrations: | ||
| + | A) 1000 cfu plated on agar plates containing chloramphenicol; | ||
| + | B) Influence of resistance induction with 1 mM IPTG, 1000 cfu where plated on agar plates containing chloramphenicol and 1mM IPTG; C) Reduced amount of <i>E.coli</i> colonies, 1000 cfu were plated on agar plates containing chloramphenicol, 1 mM IPTG to induce ampicillin resistance via BBa_K627013 and 50 µg/mL ampicillin; D) Reduced amount of <i>E.coli</i> colonies, 1000 cfu were plated on agar plates containing chloramphenicol, 1 mM IPTG to induce ampicillin resistance via BBa_K627013 and 100 µg/mL ampicillin; E) Reduced amount of <i>E.coli</i> colonies, 1000 cfu were plated on agar plates containing chloramphenicol, 1 mM IPTG to induce ampicillin resistance via BBa_K627013 and 200 µg/mL ampicillin; F) Reduced amount of <i>E.coli</i> colonies, 1000 cfu were plated on agar plates containing chloramphenicol, 1 mM IPTG to induce ampicillin resistance via BBa_K627013 and 400 µg/mL ampicillin; G) No culture growth of <i>E.coli</i> colonies, 1000 cfu were plated on agar plates containing chloramphenicol, 1 mM IPTG to induce ampicillin resistance via BBa_K627013 and 800 µg/mL ampicillin]] | ||
| - | + | This assay was performed at two temperatures. At 37°C, the optimal growth temperature and at 30°C for best protein expression conditions. All assays show the same results. The cells were able to grow at ampicilin concentrations up to 400 µg/ml. Cells transformed with the final construct containing the detection device and the protease generator device were able to survive up to 800 µg/ml ampicilin (incubated at 30°C over 2 days). | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | This leads to the conclusion, that the export of β-lactamase into the periplasm is very effective. It is also important to mention, that there is no cell survival when ampicilin is added to the media without the induction of the protease activity detection device by IPTG. | |
====Testing the "Kill Switch" by activating the protease==== | ====Testing the "Kill Switch" by activating the protease==== | ||
| - | The second major point which needs to be demonstrated is that we are able to kill the ''E.coli'' cells by activating the "Kill Switch". In the final assay we want to have the ability to induce the | + | The second major point which needs to be demonstrated is that we are able to kill the ''E.coli'' cells by activating the protease. This is referred as the "Kill Switch". In the final assay we want to have the ability to induce the protease activity detector device and the protease generate independently from each other. But first of all we have to check if our system is working and if we are able to kill the cells by activating the protease.<br> |
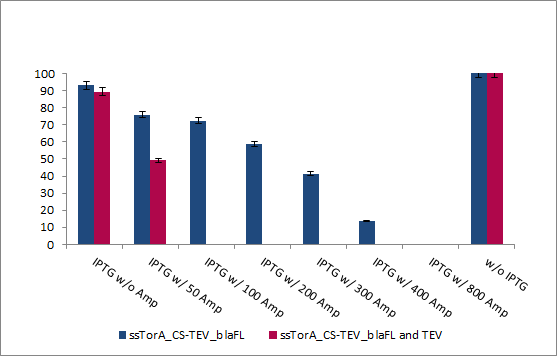
| + | To proof the principle we used the natural TEV protease and make a co-transformation with the protease activity detector device containing the cleavage site for the TEV protease. Both were induced by IPTG and so we can't control the expression rate neither the time of the induction of the protease generator device.<br> | ||
| + | After the incubation at 37°C we were able to show the desired proof of principle: when we activate the protease the resistance against ampicillin; based on the export rate of β-lactamase into the periplasm via the TAT pathway, was intimately reduced. The cells with activated TEV protease were able to survive up to concentration of 50 µg/mL ampicillin.<br> | ||
| + | [[Image:UP_bla-TEV_double-trans.png|center|500px|thumb|'''Figure 20:''' Percentage of surviving cells on different ampicillin concentration. Blue - cells transformed with the single protease activity detector device for TEV protease; Red - cell transformed with both, the protease activity detector device for TEV protease and the TEV protease itself]]<br> | ||
| + | Based on the ampicillin resistance of the plasmid of HRV14_3C protease, we were unable to show the proof of principle for this protease.<br> | ||
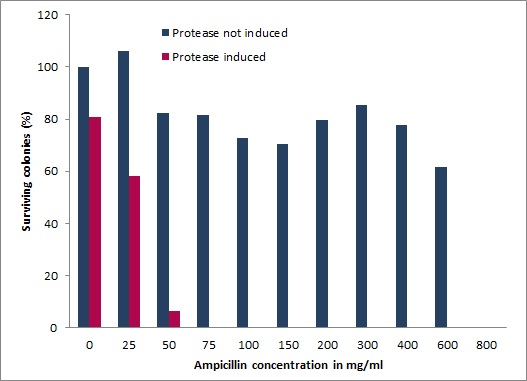
| + | The next step was to use the complete test vector which contains the protease activity detector device and the protease generator device. We introduced the arabinose inducible induction system fused to the protease flanked by the restriction sites of <i>BamH</i>I and <i>Hind</i>III into the vector of the modified Hitchhiker selection system.<br> | ||
| + | With these final vectors we had to repeat the survival screening to show that the system is also working when there is a dual induction based on only one plasmid. The cells were grown to an OD<sub>600</sub> of 0.6 and diluted to an OD<sub>600</sub> of 0.002. 100 µL of this dilution were plated on agar plates with different ampicillin concentrations and induced and induced protease. The results are shown in figure 20.<br> | ||
| + | [[Image:UP_SurvTEV_onepla.jpg|center|500px|thumb|'''Figure 21:''' Cell growth on different ampicillin concentration. Blue - cells without induction of the TEV protease; Red - cells with induced TEV protease]]<br> | ||
| + | <br> | ||
| + | The survival screening assay for TEV protease was performed 30 °C and all plates were incubated for 2 days. The ''E.coli'' XL1 blue cells were able to survive up to ampicilin concentration of 800 µg/mL, which is an increase of 100% of the original resistance capacity. When 2% arabinose is added to the media, the cells showed an intimately reduce surviving rate. The cells were able to survive at ampicilin concentrations up 25 µg/mL.<br> | ||
| + | This shows, that we got a high dynamic range from 50 up to 800 µg/mL ampicilin to screen for inhibition of the protease. | ||
| - | === | + | ===Transferring and testing the library=== |
After the successful test of the protease activity we transferred the generated <i>mdnA</i> library into our cells. The following pictures show the amazing results. Cell growth could be detected up to 400 µg Ampicillin/mL. | After the successful test of the protease activity we transferred the generated <i>mdnA</i> library into our cells. The following pictures show the amazing results. Cell growth could be detected up to 400 µg Ampicillin/mL. | ||
| - | [[Image:UP_2011-06-01_CM-Kan- | + | [[Image:UP_2011-06-01_CM-Kan-Amp_labeled02.jpg|left|420px|thumb|'''Figure 22:''' Cell growth on Ampicillin without induced protease]] |
| + | [[Image:UP_2011-06-01_CM-Kan-Ara-Amp_labeled01.jpg|right|420px|thumb|'''Figure 23:''' Cell growth on Ampicillin with induced protease]] | ||
<br><br><br><br><br><br><br><br><br><br><br><br> | <br><br><br><br><br><br><br><br><br><br><br><br> | ||
<br><br><br><br><br><br> | <br><br><br><br><br><br> | ||
| Line 95: | Line 105: | ||
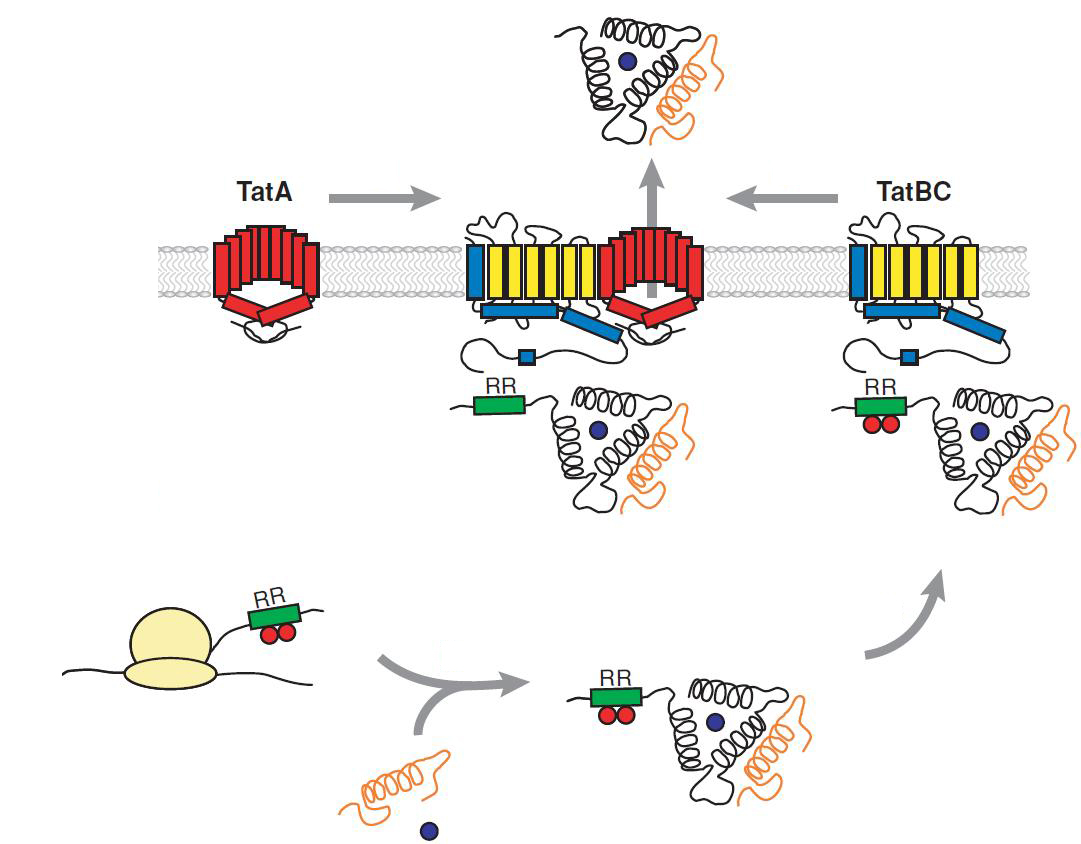
The translocon is composed of three parts: TatA, TatB and TatC. The transport-pore is proposed to be formed during substrate binding by these three parts. The TatA, TatB and TatC proteins may form complexes of different sizes, which on the other hand form pores matching the size of the folded substrate. The transport of proteins through the TAT pathway depends on the proton motive force. Calculations by Adler & Theg showed that the transport of one folded substrate molecule requires the release of approximately 7.9x10<sup>4</sup> protons, which equals 10.000 ATP molecules. The signal sequence is cleaved off after translocation. | The translocon is composed of three parts: TatA, TatB and TatC. The transport-pore is proposed to be formed during substrate binding by these three parts. The TatA, TatB and TatC proteins may form complexes of different sizes, which on the other hand form pores matching the size of the folded substrate. The transport of proteins through the TAT pathway depends on the proton motive force. Calculations by Adler & Theg showed that the transport of one folded substrate molecule requires the release of approximately 7.9x10<sup>4</sup> protons, which equals 10.000 ATP molecules. The signal sequence is cleaved off after translocation. | ||
<br> | <br> | ||
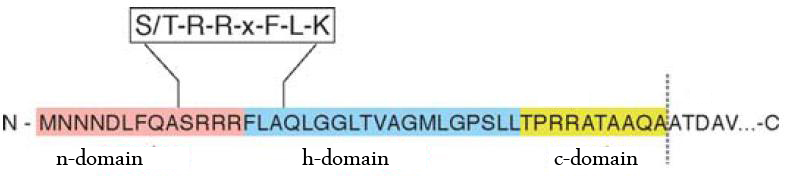
| - | [[File:UP_TorA_TATSeq2.jpg|center|450px|thumb|'''Figure | + | [[File:UP_TorA_TATSeq2.jpg|center|450px|thumb|'''Figure 24:''' Schematic representation of TorA-signal sequence. Figure adopted from: Philip A. Lee, Danielle Tullman-Ercekand George Georgiou Annu. Rev. Microbiol. 2006. 60:373–95]] |
<br> | <br> | ||
The used signal sequence originates from the TorA protein (Trimethylamin-N-Oxid-Reductase). It is the main respiratory enzyme which reduces TMAO under anaerobic conditions in the periplasm, where it is transported by the TAT pathway. | The used signal sequence originates from the TorA protein (Trimethylamin-N-Oxid-Reductase). It is the main respiratory enzyme which reduces TMAO under anaerobic conditions in the periplasm, where it is transported by the TAT pathway. | ||
<br> | <br> | ||
| - | [[File:UP_TAT_Pathway2.jpg|center|450px|thumb|'''Figure | + | [[File:UP_TAT_Pathway2.jpg|center|450px|thumb|'''Figure 25:''' Schematic representation of TAT-dependent pathway. Figure adopted from: Philip A. Lee, Danielle Tullman-Ercekand George Georgiou Annu. Rev. Microbiol. 2006. 60:373–95]] |
Latest revision as of 03:34, 29 October 2011
Contents
|
In Vivo Selection
Introduction
In addition to the phage display we developed a novel selection system. The design aimed for a inexpensive and time-saving alternative in contrast to an in vitro screen of protease inhibition. The assay allows us to select effective inhibitors for any protease, among the billions of randomly generated mutants of the Microviridin. For this purpose we designed a plasmid containing two devices, first a protease activity detector and second a protease generator. For the protease activity detector we modified the TAT-hitchhiker system developed by DeLisa et al.
We used the BioBrick <partinfo>I757010</partinfo> (β-lactamase) as well as <partinfo>K208005</partinfo> (ssTorA) and fused them together via a linker peptide. For the protease generator we cloned the arabinose induction system in front of a protease in iGEM standard.
The system works in a combined manner of the two devices. In order to confer β-lactam antibiotic resistance to cells the β-lactamase has to be exported in the periplasm. This transport is mediated by the TorA export sequence via the Twin-Arginine Translocation (TAT) system (DeLisa, 2009). The linker peptide between the TorA export sequence and the β-lactamase displays the corresponding protease cleavage site. In addition the linker peptide is chosen as short as possible to imitate folding because the TAT pathway only allows transport of correctly folded substrates. The protease as well as the linker peptide are designed as exchangeable parts.
If the protease device is functional, it will cleave the linker peptide between TorA and the β-lactamase construct which leaves the cell without any antibiotic resistance. The construct was tested with increasing ampicillin concentrations. The number of surviving colonies was depended on the export rate of the β-lactamase into the periplasm. A high cleavage rate of the linker peptide leads to a reduced ampicillin resistance. With expressed protease a dramatically drop of the number of surviving colonies could be observed.
The survival assay was carried out with and without expressed protease. By increasing of the ampicillin concentration we could detect a cutoff ampicillin concentration. The colony forming units (CFU) were counted and compared. To detected whether our constructed library is able to block the protease we co-transformed the library into E. coli carrying our pUP_SG1 plasmid.
Site-directed mutagenesis of Tobacco Etch Virus and 14_3C proteases in iGEM standard
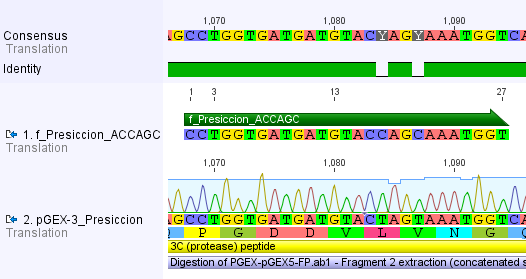
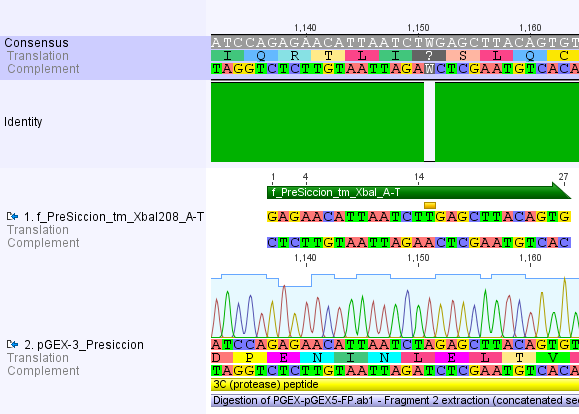
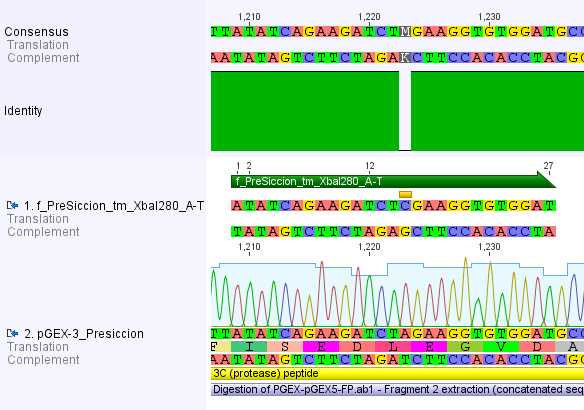
We chose two model proteases, the Tobacco Etch Virus protease, further along called TEV, and the 14_3C protease from the human rhinovirus, also known as PreScission. Both proteases contained iGEM restriction sites, one in case of the TEV and three for 14 3C, respectively.
Thus, site-directed mutagenesis was applied to mutate each restriction site in E. coli. For every mutation a forward and a reverse primer had to be designed. The following pictures show the forward primers for the side directed mutagenesis, the reverse primers are the reverse complement sequence of the forward primers.
For the 14_3C protease:
For the TEV protease:
We used assembly PCR to gain the final product. In case of 14_3C two assembly PCRs had to be done. The expected size of the fragments are shown in the gel picture. In the following picture all fragments contain the correct sizes.
Construction of two component for the in vivo selection assay
The in vivo assay is based on two devices. On the one hand the protease activity detector device, on the other a protease generator device. The detector device is based on the enzyme β-lactamase, which is only active inside the periplasm and provides the resistance against lactam antibiotics like ampicillin and penicillin. For the protease generator device we needed a second induction system on our plasmid, which is quite uncommon for any plasmid. So several problems had to solved to set up an effective in vivo selection assay:
- a second, very tight induction system has to be found
- a way to control the export rate of β-lactamase into the periplasm, which can be reduced by the protease
We solved these problems by putting the protease detector device under the control of the lac-operon. The protease device is induced by the arabinose system of the pBAD_iGEMexpress vector. The idea to control the survival of the cells via the amount of exported protein is inspired by the Hitchhiker assay of DeLisa et al. Using the TAT pathway for the export of β-lactamase into the periplasm allows us to introduce a protease specific cleavage site in front of the signal sequence regulating the export.
Introducing the specific protease cleavage site
To adapt the protease activity detector device to any protease we want to screen, we had to modularize it. Our detector device was set up on a vector for the Hitchhiker selection. The plasmid offers two unique restriction sites on the plasmid, after the signal sequence of TorA one restriction site for XhoI and a NheI restriction site in front of the β-lactamase. Both were used to introduce the specific protease cleavage site in between the TorA signal sequence and the β-lactamase. By designing two complementary oligonucleotides containing the specific protease cleavage site flanked by two short linker regions. After the digest of the vector with XhoI and NheI the hybridized oligonucleotides, containing the overhangs for XhoI and NheI can be ligated into the vector.
Adding the arabinose inducible induction system to control the proteases, TEV and 14_3C
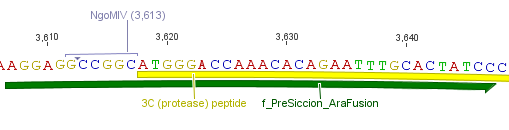
We needed a time independent induction of the protease device. Therefore we cloned the arabionse induction system (araC) in front of the protease device out of the pBAD_iGEMexpress vector. The fusion of gene construct and induction system was always performed via an NgoMIV restriction site. This restriction site was introduced with our amplification primers.
We used the RFC 23 cloning strategy to get a fusion protein of our desired protease and the induction system, which allows us to induce the protease. Both genes were amplified with PCR using two special primers generating a NgoMIV restrictioin site at the end of the induction system and in front of the protease. The amplified protease were digested with BamHI and NgoMIV, the arabinose inducible induction system with NgoMIV and HindIII. The vector, which contains the ssTorA_CS-Protease_β-lactamase device was digested with HindIII and BamHI. A triple ligation yielded the final construct with both devices: the protease detector and the protease generator. The two pictures below show the final vector constructs , which where used for the survival screening.
Investigation of the in vivo selection assay
Two major points have to be demonstrated: First, the export of β-lactamase over the TAT pathway has to be efficient enough to provide a certain resistance against ampicillin and second, the activated protease is able to intimately reduce the resistance against ampicillin by cleavage of the TorA signal sequence from the β-lactamase.
Testing the export efficiency of the protease activity detector
To test the export rate of the β-lactamse of the protease activity detector device we grew an E. coli culture up to an OD600 of 0.6. The cells were then diluted to an OD of 0.002 and 100 µL were plated in triplicates with increasing ampicillin concentrations. The graph below shows the amount of surviving cells in percentage.
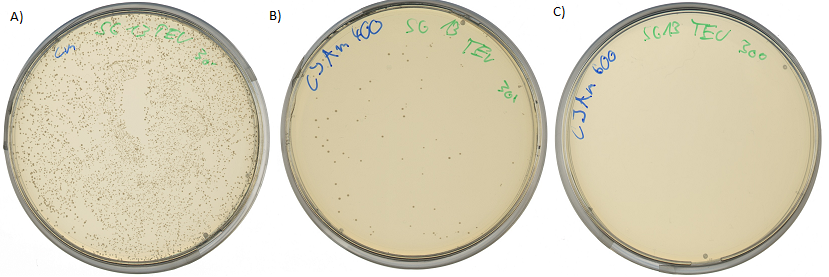
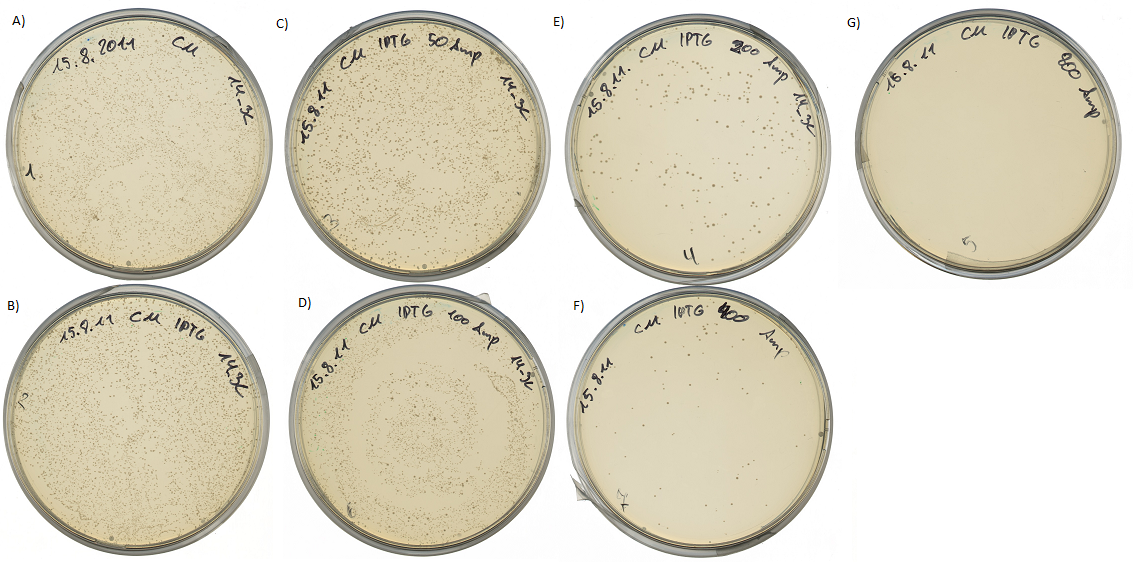
This assay was performed at two temperatures. At 37°C, the optimal growth temperature and at 30°C for best protein expression conditions. All assays show the same results. The cells were able to grow at ampicilin concentrations up to 400 µg/ml. Cells transformed with the final construct containing the detection device and the protease generator device were able to survive up to 800 µg/ml ampicilin (incubated at 30°C over 2 days).
This leads to the conclusion, that the export of β-lactamase into the periplasm is very effective. It is also important to mention, that there is no cell survival when ampicilin is added to the media without the induction of the protease activity detection device by IPTG.
Testing the "Kill Switch" by activating the protease
The second major point which needs to be demonstrated is that we are able to kill the E.coli cells by activating the protease. This is referred as the "Kill Switch". In the final assay we want to have the ability to induce the protease activity detector device and the protease generate independently from each other. But first of all we have to check if our system is working and if we are able to kill the cells by activating the protease.
To proof the principle we used the natural TEV protease and make a co-transformation with the protease activity detector device containing the cleavage site for the TEV protease. Both were induced by IPTG and so we can't control the expression rate neither the time of the induction of the protease generator device.
After the incubation at 37°C we were able to show the desired proof of principle: when we activate the protease the resistance against ampicillin; based on the export rate of β-lactamase into the periplasm via the TAT pathway, was intimately reduced. The cells with activated TEV protease were able to survive up to concentration of 50 µg/mL ampicillin.
Based on the ampicillin resistance of the plasmid of HRV14_3C protease, we were unable to show the proof of principle for this protease.
The next step was to use the complete test vector which contains the protease activity detector device and the protease generator device. We introduced the arabinose inducible induction system fused to the protease flanked by the restriction sites of BamHI and HindIII into the vector of the modified Hitchhiker selection system.
With these final vectors we had to repeat the survival screening to show that the system is also working when there is a dual induction based on only one plasmid. The cells were grown to an OD600 of 0.6 and diluted to an OD600 of 0.002. 100 µL of this dilution were plated on agar plates with different ampicillin concentrations and induced and induced protease. The results are shown in figure 20.
The survival screening assay for TEV protease was performed 30 °C and all plates were incubated for 2 days. The E.coli XL1 blue cells were able to survive up to ampicilin concentration of 800 µg/mL, which is an increase of 100% of the original resistance capacity. When 2% arabinose is added to the media, the cells showed an intimately reduce surviving rate. The cells were able to survive at ampicilin concentrations up 25 µg/mL.
This shows, that we got a high dynamic range from 50 up to 800 µg/mL ampicilin to screen for inhibition of the protease.
Transferring and testing the library
After the successful test of the protease activity we transferred the generated mdnA library into our cells. The following pictures show the amazing results. Cell growth could be detected up to 400 µg Ampicillin/mL.
Background
Twin Arginine Translocon (TAT)
The TAT pathway is responsible for the transport of folded proteins across energy-transducing membranes and it is able of discriminating of unfolded proteins. This transport pathway is common for bacteria, archea and plants and translocates circa 6% of E.coli produced secreted proteins. The translocated proteins must have a signal peptide for targeting of the TAT-transporter, such proteins are e.g. hydrogenases, dehydrogenases and reductases. The signal sequence itself is composed of an N-terminal positive charged domain, a hydrophobic domain and a C-terminal domain.
The translocon is composed of three parts: TatA, TatB and TatC. The transport-pore is proposed to be formed during substrate binding by these three parts. The TatA, TatB and TatC proteins may form complexes of different sizes, which on the other hand form pores matching the size of the folded substrate. The transport of proteins through the TAT pathway depends on the proton motive force. Calculations by Adler & Theg showed that the transport of one folded substrate molecule requires the release of approximately 7.9x104 protons, which equals 10.000 ATP molecules. The signal sequence is cleaved off after translocation.
The used signal sequence originates from the TorA protein (Trimethylamin-N-Oxid-Reductase). It is the main respiratory enzyme which reduces TMAO under anaerobic conditions in the periplasm, where it is transported by the TAT pathway.
Tobacco Etch Virus (TEV) protease
TEV protease is the common name for the 27 kDa catalytic domain of the Nuclear Inclusion a endopeptidase (NIa) encoded by the tobacco etch virus. TEV protease is a useful reagent for cleaving fusion proteins. It recognizes a linear epitope of the general form E-Xaa-Xaa-Y -Xaa-Q-(G/S), with cleavage occurring between Q and G or Q and S. In TEV protease the serine nucleophile of the conventional Ser-Asp-His triad is a cysteine instead. This probably explains why TEV protease is resistant to many commonly used protease inhibitors.
14_3C-Protease
The 14_3C protease originates from the human rhinovirus. Rhinoviruses are the most frequent reason for infections of the upper and lower respiratory tract, also known as the cold. Because the 3C protease of human rhinovirus is necessary for the cleavage of the polyprotein translated from viral RNA it may serve as a potential target for development of antiviral targets. The recombinant type 14_3C protease from human rhinovirus (HRV 3C) recognizes the same cleavage site as the native enzyme: LeuGluValLeuPheGln↓GlyPro. The small, 22-kDa size of the protease got its optimal activity at 4°C but is still very active at 37 °C. It is commonly used for an easy tag removal after the purification of recombinant proteins carrying his-tag. The 14_3C works with a catalytic triade, containing the amino acid residues Ser-Asp-His at its active site.
References
- Cabrita, L. D., Gilis, D., Robertson, A. L., Dehouck, Y., Rooman, M. and Bottomley, S.P. (2007) Enhancing the stability and solubility of TEV protease using in silico design. Protein Sci. 16: 2360-2367
- Genest O., Ilbert M., Méjean V., Iobbi-Nivol C.,(2005) TorD, an Essential Chaperone for TorA Molybdoenzyme Maturation at High Temperature. J. Biol. Chem. 280: 15644-15648.
- Kapust R. B., Tözsér J., Fox J. D., Anderson D. E., Cherry S., Copeland T. D., Waugh D. S. (2001) Tobacco etch virus protease: mechanism of autolysis and rational design of stable mutants with wild-type catalytic proficiency. Protein Eng. 12:993-1000.
- Lee P. A., Tullman-Ercekand D., Georgiou G., (2006) The bacterial twin-arginine translocation pathway. Annu. Rev. Microbiol. 60:373–95
- Lucast, L. J., Batey, R. T., and Doudna, J. A. (2001). Large-scale purification of a stable form of recombinant tobacco etch virus protease. Biotechniques 30:544-550.
- Shih S. R., Chen S. J., Hakimelahi G. H., Liu H. J., Tseng C. T., Shia K. S., (2004) Selective human enterovirus and rhinovirus inhibitors: An overview of capsid-binding and protease-inhibiting molecules. Med Res Rev 24(4):449-74.
- Wanga QM, Chen SH., (2007) Human rhinovirus 3C protease as a potential target for the development of antiviral agents. Curr Protein Pept Sci. 8(1):19-27.
- Waraho D., DeLisa M. P., (2009). Versatile selection technology for intracellular protein-protein interactions mediated by a unique bacterial hitchhiker transport mechanism. PNAS 106(10):3692-7
 "
"