Team:BU Wellesley Software/Notebook/VanessaNotebook
From 2011.igem.org
| (73 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | <html> | ||
| + | <head> | ||
| + | <title>BU-Wellesley iGEM Team: Welcome</title> | ||
| + | <meta http-equiv="Content-Type" content="text/html; charset=utf-8"> | ||
| + | <script src="http://cdn.jquerytools.org/1.2.5/full/jquery.tools.min.js?foo"></script> | ||
| + | <style type="text/css"> | ||
| + | /*hide default igem banner and reformat style into blank slate*/ | ||
| + | #globalWrapper {width: 100%;} | ||
| + | #top-section {width: 100%; height:30px; border:none;} | ||
| + | #p-logo {display:none;} | ||
| + | #search-controls {display:none;} | ||
| + | #menubar a {color:#000000;} | ||
| + | #menubar a:hover{text-decoration:none; color:#52749C;} | ||
| + | .left-menu {background-color:#FFFFFF; margin:5px 0px 0px 0px; padding:0;} | ||
| + | .left-menu ul {background-color:#FFFFFF; margin:0; padding:0;} | ||
| + | .right-menu ul li a {background-color:#FFFFFF;} | ||
| + | .printfooter {display:none;} | ||
| + | #footer-box {border:none;} | ||
| + | #catlinks {display:none;} | ||
| + | .firstHeading {display:none;} | ||
| + | #content {width: 100%; border:none;} | ||
| + | #bodyContent {border:none;} | ||
| + | |||
| + | /*actual content styles*/ | ||
| + | body {width: 800px; margin:auto;} | ||
| + | |||
| + | #bu-wellesley_wiki_content {height:auto; line-height:100%;} | ||
| + | /*#bu-wellesley_wiki_content a {color:#69d01d;}*/ | ||
| + | #bu-wellesley_wiki_content a:hover {text-decoration:none; color:#3d3f3c;} | ||
| + | |||
| + | .navbar li {color: #ffffff;} | ||
| + | .navbar li a {color: #ffffff;} | ||
| + | .navbar li a:hover {background:#69d01d; color: #ffffff;} | ||
| + | |||
| + | /*only use for current page content header (i.e. Team, G-nomeSurferPro, etc)*/ | ||
| + | H6 { | ||
| + | font-family: Helvetica; | ||
| + | text-transform: uppercase; | ||
| + | text-decoration: none; | ||
| + | text-align: left; | ||
| + | color: #3d3f3c; | ||
| + | font-size: 32pt; | ||
| + | } | ||
| + | |||
| + | </style> | ||
| + | |||
| + | <link rel="stylesheet" type="text/css" href="http://cs.wellesley.edu/~hcilab/iGEM_wiki/css/Team.css"> | ||
| + | |||
| + | </head> | ||
| + | <body class="basiclayout"> | ||
| + | <div id="bu-wellesley_wiki_content"> | ||
| + | |||
| + | <p style="text-align:center;"><a href="https://2011.igem.org/Team:BU_Wellesley_Software"><img src="http://cs.wellesley.edu/~hcilab/iGEM_wiki/images/banner.png" width="800px"></a></p> | ||
| + | |||
| + | <ul id="nav"> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Team">Team</a></li> | ||
| + | <li><a href="#">Project</a> | ||
| + | <ul> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Project_Overview">Overview</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Clotho">Clotho</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/G-nomeSurferPro">G-nome Surfer Pro</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/OptimusPrimer">Optimus Primer</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Trumpet">Trumpet</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Puppetshow">Puppetshow</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/eLabNotebook">eLabNotebook</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Wet_Lab">Wet Lab</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Downloads_and_Tutorials">Downloads and Tutorials</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="#">Process</a> | ||
| + | <ul> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Methodology">Methodology</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Safety">Safety</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Notebook">Notebook</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Outreach">Outreach</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Tips">Tips and Tricks</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Gold">Medal Fulfillment</a></li> | ||
| + | <li><a href="#">Additional Info</a> | ||
| + | <ul> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Acknowledgement">Acknowledgement</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Social">Fun</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | </ul> | ||
| + | |||
| + | <br> | ||
| + | <h6>Vanessa's Notebook</h6> | ||
| + | </body> | ||
| + | </html> | ||
| + | |||
| + | |||
| + | |||
| + | *[[Lab Protocols | Protocols]] | ||
| + | |||
__TOC__ | __TOC__ | ||
| - | ==Week 1: 6/1-6/10 | + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#333333;margin-top:5px;" cellspacing="18" |
| + | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h3>Week 1: 6/1-6/10</h3> | ||
| + | |- | ||
| + | | | ||
| + | |||
The first few days, the group sat through a boot camp, where we learned about biobricks, the basics of Biology such as DNA transcription and translation, and how to use Clotho tools. As exposure to the more computer science aspect of the project, the group learned how to write a Clotho application. | The first few days, the group sat through a boot camp, where we learned about biobricks, the basics of Biology such as DNA transcription and translation, and how to use Clotho tools. As exposure to the more computer science aspect of the project, the group learned how to write a Clotho application. | ||
| Line 8: | Line 108: | ||
The transformations produced several cultures that were both dark and light in color. Plasmid preps were made of the dark and light cultures of the UV promoter. The dark culture had an okay amount of DNA concentration. However the other samples did not have a good concentration of DNA either. | The transformations produced several cultures that were both dark and light in color. Plasmid preps were made of the dark and light cultures of the UV promoter. The dark culture had an okay amount of DNA concentration. However the other samples did not have a good concentration of DNA either. | ||
| - | There seemed to be contamination or something else growing on the plates because some of the cultures were oddly shaped and there was a pungent smell. As a result, a sample of plasmid prep culture was stained and looked under the microscope. Most samples did not have E. coli in the sample. | + | There seemed to be contamination or something else growing on the plates because some of the cultures were oddly shaped and there was a pungent smell. As a result, a sample of plasmid prep culture was stained and looked under the microscope. Most samples did not have E. coli in the sample. |
| + | |} | ||
| - | ==Week 2: 6/11-6/17 | + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#333333;margin-top:5px;" cellspacing="18" |
| + | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h3>Week 2: 6/11-6/17</h3> | ||
| + | |- | ||
| + | | | ||
A second round of plasmid preps were made of the same plates to troubleshoot the cultures that do not have high concentrations of DNA. These unfortunately still did not work or produce anything. Only the UV promoter sample had a culture. | A second round of plasmid preps were made of the same plates to troubleshoot the cultures that do not have high concentrations of DNA. These unfortunately still did not work or produce anything. Only the UV promoter sample had a culture. | ||
| Line 21: | Line 125: | ||
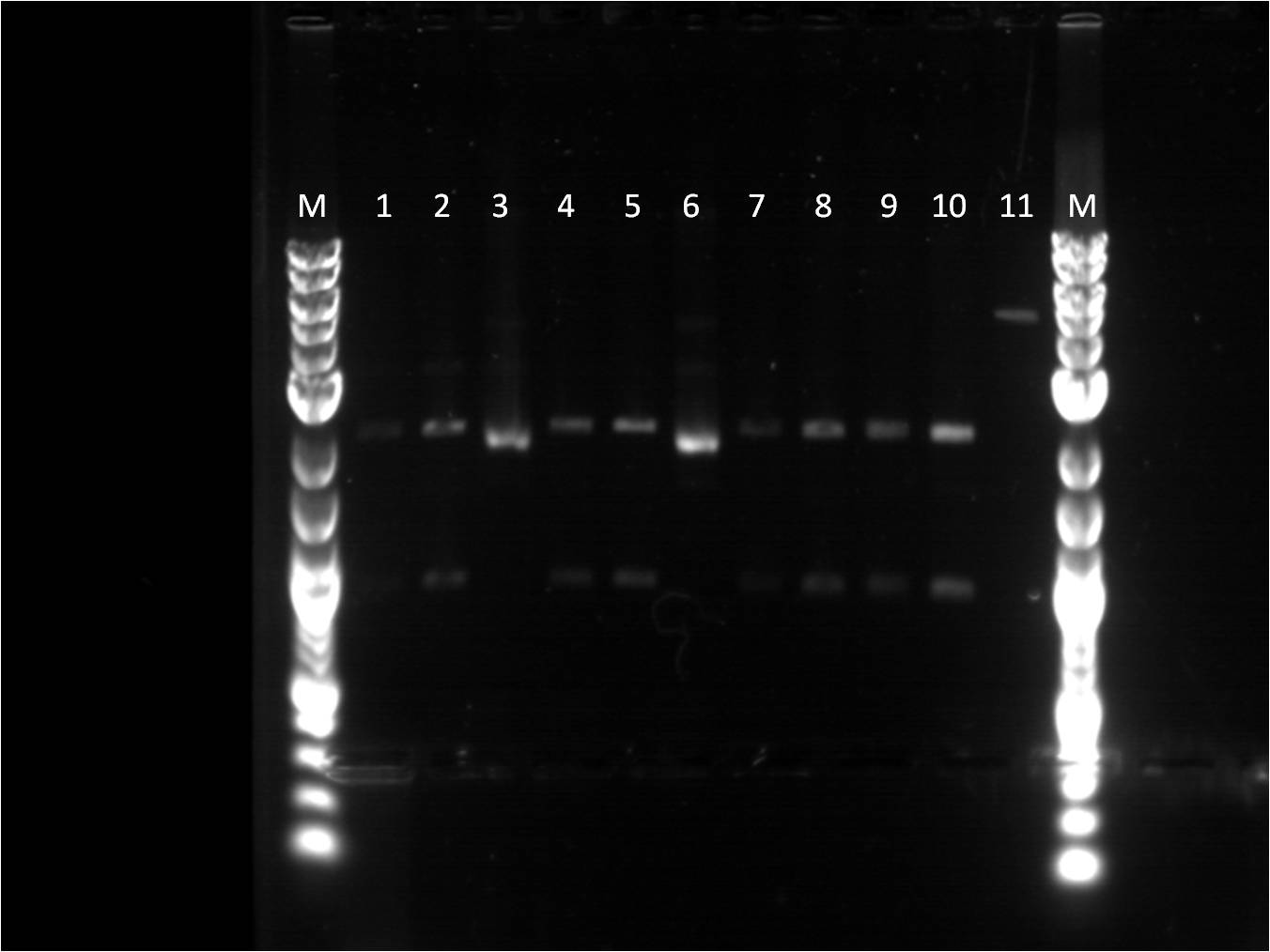
New transformations were made of the 7 original parts chosen and few additional ones (RBS, promoters, RFP). Most of these seem to grow successfully, however 2 plates had cultures with a pink tint to them. There was one plate where nothing grew on it. There was also two negative controls for ampicillin plates and kanamycin plates, where nothing grew. Plasmid preps of the new transformations were made. | New transformations were made of the 7 original parts chosen and few additional ones (RBS, promoters, RFP). Most of these seem to grow successfully, however 2 plates had cultures with a pink tint to them. There was one plate where nothing grew on it. There was also two negative controls for ampicillin plates and kanamycin plates, where nothing grew. Plasmid preps of the new transformations were made. | ||
| - | [[Image: | + | [[Image: 6_17_2011_miniprep_labeled.jpeg | left | 300px |thumb| This is a figure of a gel with several samples for verification.]] |
| - | ==Week 3: 6/18-6/24 | + | |
| + | |||
| + | |||
| + | |||
| + | Wells 1 through 9 and M being the location of the | ||
| + | ladder. | ||
| + | 1. Bba_I13453 18.6 ng/uL | ||
| + | 2. Bba_J23100 53 ng/uL | ||
| + | 3. Bba_J61127 23.1 ng/uL | ||
| + | 4. Bba_E1010 56.5 ng/uL | ||
| + | 5. Bba_J23101 32.6 ng/uL | ||
| + | 6. Bba_14053 17.5 ng/uL | ||
| + | 7. Bba_J23109 31.6 ng/uL | ||
| + | 8. Bba_J61101 9.6 ng/uL | ||
| + | 9. Bba_J61100 30.2 ng/uL | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |} | ||
| + | |||
| + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#333333;margin-top:5px;" cellspacing="18" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h3>Week 3: 6/18-6/24</h3> | ||
| + | |- | ||
| + | | | ||
- Prepared plasmid preps and continued to used the QIAcube for minipreps of biobrick parts listed below: | - Prepared plasmid preps and continued to used the QIAcube for minipreps of biobrick parts listed below: | ||
| Line 54: | Line 187: | ||
Two approaches were decided on to make the 4 different successful devices, the Bottom Up approach and using composite parts with flourescent proteins to build the some of the devices. The team broke up into two. I became part of the team, where we use the composite parts approach using YFP, GFP, and RFP. I am responsible for making a device with the yellow fluorescent protein composite parts along with four different promoters found in biobrick parts. The promoters include Bba_I13453 (pbad), Bba_I14033 (pcat), Bba_R0040 (ptet), and Bba_R2000. | Two approaches were decided on to make the 4 different successful devices, the Bottom Up approach and using composite parts with flourescent proteins to build the some of the devices. The team broke up into two. I became part of the team, where we use the composite parts approach using YFP, GFP, and RFP. I am responsible for making a device with the yellow fluorescent protein composite parts along with four different promoters found in biobrick parts. The promoters include Bba_I13453 (pbad), Bba_I14033 (pcat), Bba_R0040 (ptet), and Bba_R2000. | ||
| + | |||
To Prepare for Ligation of device: | To Prepare for Ligation of device: | ||
| Line 60: | Line 194: | ||
- Restriction digest, gel run and extraction with Bba_I13453 (pbad), Bba_I14033 (pcat), and Bba_E0340 (YFPc) | - Restriction digest, gel run and extraction with Bba_I13453 (pbad), Bba_I14033 (pcat), and Bba_E0340 (YFPc) | ||
| + | - Restriction Digest: Bba_I13453 cut with SpeI and PstI | ||
| - | |||
| - | |||
Ligation: | Ligation: | ||
| - | Five samples were made at 3:1 ratio of insert to backbone. | + | Five samples were made at 3:1 ratio of insert to backbone. |
| - | + | - pcat + YFPc | |
| - | + | - pcat + YFPc | |
| - | + | - pbad + YFPc | |
| - | + | - pbad + YFPc | |
| - | + | - pbad + YFPc | |
Transformations of these samples were done, but unfortunately nothing grew on the plates. | Transformations of these samples were done, but unfortunately nothing grew on the plates. | ||
| + | |} | ||
| + | |||
| + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#333333;margin-top:5px;" cellspacing="18" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h3>Week 4: 6/25-7/1</h3> | ||
| + | |- | ||
| + | | | ||
| + | |||
| + | |||
| + | - Restriction Digest 3 samples of YFPc with Xbal and PstI and R2000 with SpeI and PstI. | ||
| + | |||
| + | [[Image: 6_28_11_RD_labeled.jpeg | left | 350px |thumb| Gel of Restriction Digest for multiple samples of YFPc and a R 2000.]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | 1. Restriction digest of Bba_E0430.1 (YFPc) | ||
| + | 2. " " " " | ||
| + | 3. Uncut Bba_E0430.1 (YFPc) | ||
| + | 4. Restriction digest of Bba_E0430.2 (YFPc) | ||
| + | 5. " " " " | ||
| + | 6. Uncut Bba_E0430.2 (YFPc) | ||
| + | 7. Restriction digest of Bba_E0430.2 (YFPc) | ||
| + | 8. " " " " | ||
| + | 9. Restriction digest of Bba_E0430 (YFPc) | ||
| + | 10. " " " " | ||
| + | 11. Restriction digest of Bba_R2000 | ||
| + | |||
| - | |||
| - | |||
- These samples ran on a gel, extracted, and quantified. Gel quantification values were ranging in the 20s for the samples listed above. | - These samples ran on a gel, extracted, and quantified. Gel quantification values were ranging in the 20s for the samples listed above. | ||
| + | |||
| + | |||
| + | |||
- Restriction Digest of Bba_I14033 (pcat), Bba_I13453 (pbad), Bba_R2000, Bba_E0240 (GFPc), and Bba_R0040. | - Restriction Digest of Bba_I14033 (pcat), Bba_I13453 (pbad), Bba_R2000, Bba_E0240 (GFPc), and Bba_R0040. | ||
| Line 88: | Line 249: | ||
- Ligation: | - Ligation: | ||
| - | + | Five samples were ligated at a 6:1 ratio of insert to backbone. | |
| - | + | - Bba_R2000 + Bba_E0430 | |
| - | + | - Bba_I14033 + Bba_E0430 | |
| - | + | - Bba_R0040 + E0430 | |
| - | + | - Bba_R2000.1.2 + E0430 | |
| - | + | - Bba_I13453 + E0430 | |
:. These 5 ligation samples were also transformed, but nothing grew after incubating them overnight at 37 degrees Celsius. | :. These 5 ligation samples were also transformed, but nothing grew after incubating them overnight at 37 degrees Celsius. | ||
| - | A new | + | |
| + | A new ligation protocol was attempted for two new samples. The ligation samples are Bba_R2000 + Bba_E0430 with one sample being a 3:1 insert to backbone and the other 6:1. The samples will be held at 16 degrees Celsius for 12 hours and holding it at 4 degrees Celsius for 4 hours. | ||
| + | |} | ||
| + | |||
| + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#333333;margin-top:5px;" cellspacing="18" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h3>Week 5: 7/2-7/8</h3> | ||
| + | |- | ||
| + | | | ||
| + | |||
| + | |||
| + | The new ligation samples were transformed over the weekend on plates. On Tuesday morning, there was growth on the plates; however, there weren't any colonies growing, just a lawn of bacteria. It was concluded that the plates may have not had the antibiotic to be able to grow the specific parts that were ligated. Another attempt was made, but as plasmid preps were made it was discovered the LB broth had some contamination. This added to the transformation problem because the same LB broth was used to plate the samples. Because of contamination, new LB broth was made as well as new plates with the appropriate antibiotics. | ||
| + | |||
| + | Another attempt to transform last week's ligation samples, but unfortunately again no growth. | ||
| + | |||
| + | While the ligation issues are being figured out, stocks of parts that are needed for future ligations are being made. | ||
| + | |||
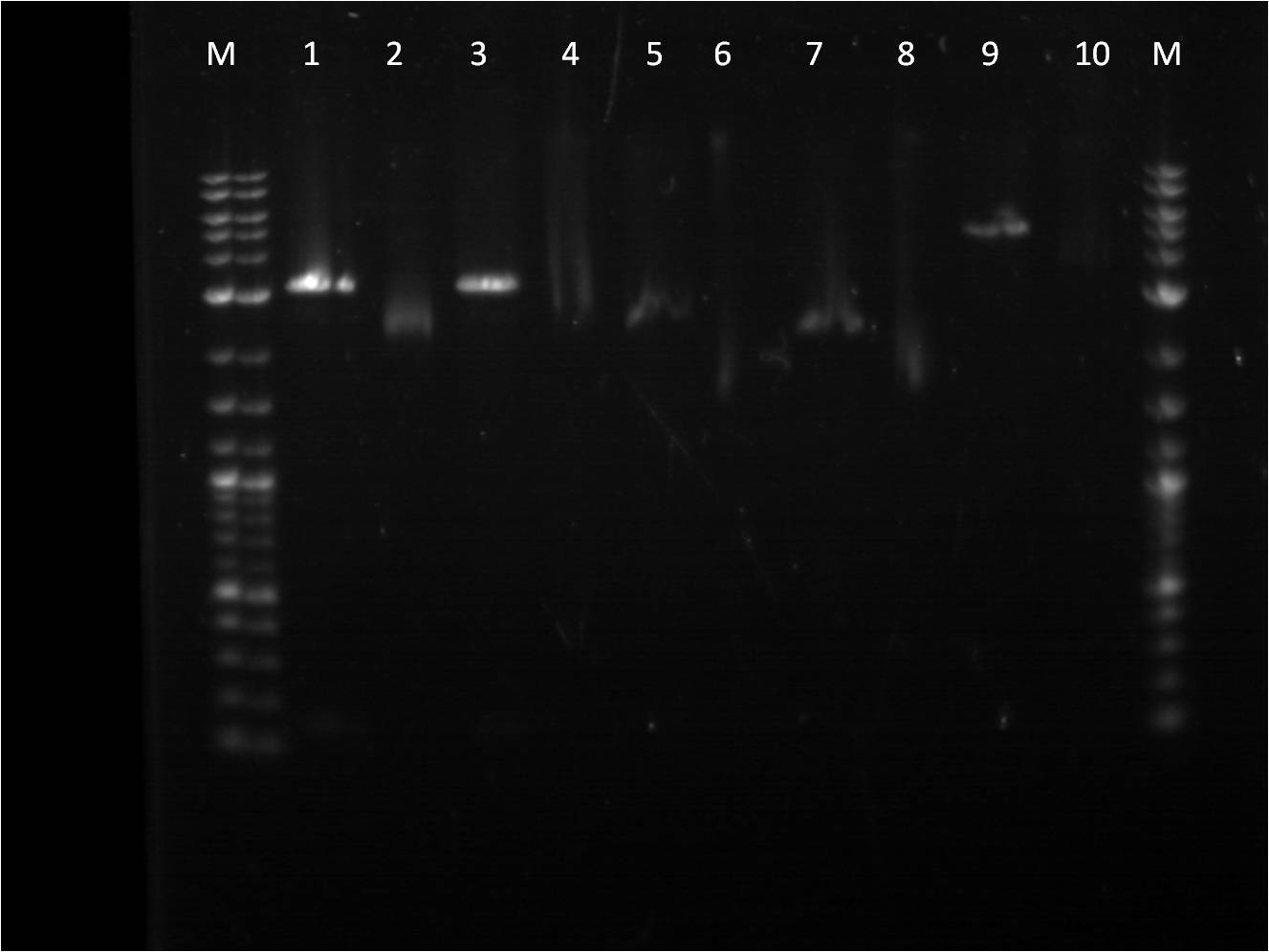
| + | Three promoters needed in gel extraction stage are promoters, Bba_I13453, Bba_R0040, and Bba_I14033. These biobrick parts were cut with SpeI and PstI using a higher mass of DNA. | ||
| + | |||
| + | These pieces were run on a gel providing some strange unclear results on the image below. | ||
| + | |||
| + | [[Image: 7_5_11_RD_VYKJ_labeled.jpeg | cleft | 300px |thumb| This is a figure of a gel with restriction digest samples.]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | Wells: | ||
| + | 5. Restriction Digest of Bba_I13453.2 | ||
| + | 6. Uncut Bba_I13453.2 | ||
| + | 7. Restriction Digest of Bba_R0040 | ||
| + | 8. Uncut Bba_R0040 | ||
| + | 9. Restriction Digest of Bba_I14033 | ||
| + | 10.Uncut Bba_I14033 | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | - Only the well 5 and 7 were cut and quantified. | ||
| + | |||
| + | - Plasmid preps of various parts: Bba_E0430, Bba_I14033, Bba_I13453, Bba_E0240 | ||
| + | |||
| + | - Streaked 3 plates from -20 degrees Celsius stock (Bba_R2000, Bba_R0040, and Bba_I13453) | ||
| + | |} | ||
| + | |||
| + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#333333;margin-top:5px;" cellspacing="18" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h3>Week 6: 7/9-7/15</h3> | ||
| + | |- | ||
| + | | | ||
| + | |||
| + | - Visited MIT | ||
| + | |||
| + | - A few of the team members went to talk to MIT's postdoc, Jonathan to troubleshoot our ligation and transformation issues. He suggested checking our gel extraction samples, using a different transformation and ligation protocol. | ||
| + | |||
| + | - We thought there may be an issue with out TOP10 competent cells. As a result, we transformed ligation reactions in TOP10 competent cells and commercial cells to see if that was affecting our results. We noticed that once the cells were spun down the pellet of cells was much larger for the commercial cells as opposed to the TOP10 cells made by us. Unfortunately, nothing grew on the plates. | ||
| + | |||
| + | - Ligation reactions were attempted again between the YFP composite and various promoters. | ||
| + | |||
| + | - Since we thought that the backbones might be re-ligating with itself, we decided to use CIP on the backbone, which dephosphorylates the backbone. These reactions/samples had to be run through a gel, extracted, and quantified. | ||
| + | |||
| + | - Ligation reactions were made with CIP on the backbone and without CIP, using quick ligase. These reactions were transformed onto plates, but unfortunately did not grow. | ||
| + | |||
| + | |} | ||
| + | |||
| + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#333333;margin-top:5px;" cellspacing="18" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h3>Week 7: 7/16-7/22</h3> | ||
| + | |- | ||
| + | | | ||
| + | |||
| + | As a team, we brainstormed ideas on how to find the right combination for ligation protocol between reaction time and ligation ratios. | ||
| + | |||
| + | - I tried ligation reactions with Bba_E0430(YFPc) and Bba_R0040(ptet) for 30 minutes, 1 hour, and overnight on the bench. The reactions were the same sample with two different ratios 1:3 and 1:7. I transformed these reactions with TOP10 competent cells. Unfortunately, nothing grew on the plates this try. With the 1:3 ratio, I tried transforming with 4 tubes of TOP10 competent cells, but nothing worked either. | ||
| + | |||
| + | - The ligation reaction was transformed with alpha-select competent cells and there was success. A few colonies grew on each plate with various reactions. It grew best at 1:3 for 30 minutes reaction time on bench at room temperature. To confirm these results, 4 plasmid preps were taken from each plate. There were a few with false positives, but the others seem to be at the right position on the gel with okay quantification values. | ||
| + | |||
| + | - Two other constructs were ligated and checked on a gel to see if it fits right with what was needed. Unfortunately, the ligation of YFPc and R2000 did not show up on the gel, there must be some kind of error in the reaction. | ||
| + | |} | ||
| + | |||
| + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#333333;margin-top:5px;" cellspacing="18" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h3>Week 8: 7/23-7/29</h3> | ||
| + | |- | ||
| + | | | ||
| + | |||
| + | Because our ligations seemed to be working successfully, we began to restriction map the various constructs.Before this, the plates with various constructs were checked under UV light. The construct with the constitutive promoter, pcat, BBa_I14033 did not glow under the UV light; however, the construct with ptet_YFPc (BBa_R0040_BBa_E0430) did glow brightly under the UV light. The other constitutive promoter, BBa_R2000 made BBa_E0240 glow green under the UV light. The inducible promoter, pbad (BBa_I13453), had the correct response in which it did not glow because there was no arabinose present. In addition, the constructs were checked by making 4 plasmid preps of each plate, just in case there were false positives on the plate. The plasmids of each plate were then ran on a gel. The constructs tested include: | ||
| + | |||
| + | [[Image: 7_25_11_KJ_rd_VY_ligations.png | right | 370px |thumb| This is a figure of a gel with construct samples.]] | ||
| + | |||
| + | |||
| + | |||
| + | 12-15.BBa_I13453_BBa_E0430 (Pbad_YFPc) | ||
| + | 16-19.BBa_I14033_BBa_E0430 (Pcat_YFPc) | ||
| + | 20.BBa_R2000_BBa_E0430 (promoter_YFPc) | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | Also, one of the constructs with promoter pbad is an inducible promoter. Therefore, this construct must be induced with arabinose so it can express the colors, green, red, and in my case yellow. We attempted to induce this promoter by a concentration of arabinose of 50 uM.Unfortunately this did not cause the cell culture to glow. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | Once the plasmids checked out, it was decided to restriction map all of the constructs just to verify the correct parts were ligated. The construct, pbad_YFPc and pcat_YFPc had a single cut and double cut to check the insert and the whole plasmid size. BBa_I14033_BBa_E0240 (pcat_GFPc) was also cut twice to check the various sizes. The products of the restrcition digest was run on a gel. | ||
| + | |||
| + | |||
| + | [[Image: 7_29_11_RD_AT_VY.png | left | 350px |thumb| This is a figure of a gel with multiple cut construct samples.]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | 7-9: BBa_I13453_BBa_E0430 (pbad_YFPc); uncut, cut, double cut (It has the correct size bands) | ||
| + | 10-12: BBa_I14033_BBa_E0430 (pcat_YFPc); uncut, cut, double cut (It has incorrect size bands and | ||
| + | well 12 does not have two bands. It seems that a double cut was not made.) | ||
| + | 13-15: BBa_I14033_BBa_E0240 (pcat_GFPc) uncut, cut, double cut (These bands also seem wrong | ||
| + | according to the DNA ladder. The bands for the double cut seem too high) | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |} | ||
| + | |||
| + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#333333;margin-top:5px;" cellspacing="18" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h3>Week 9: 7/30-8/5</h3> | ||
| + | |- | ||
| + | | | ||
| + | |||
| + | |||
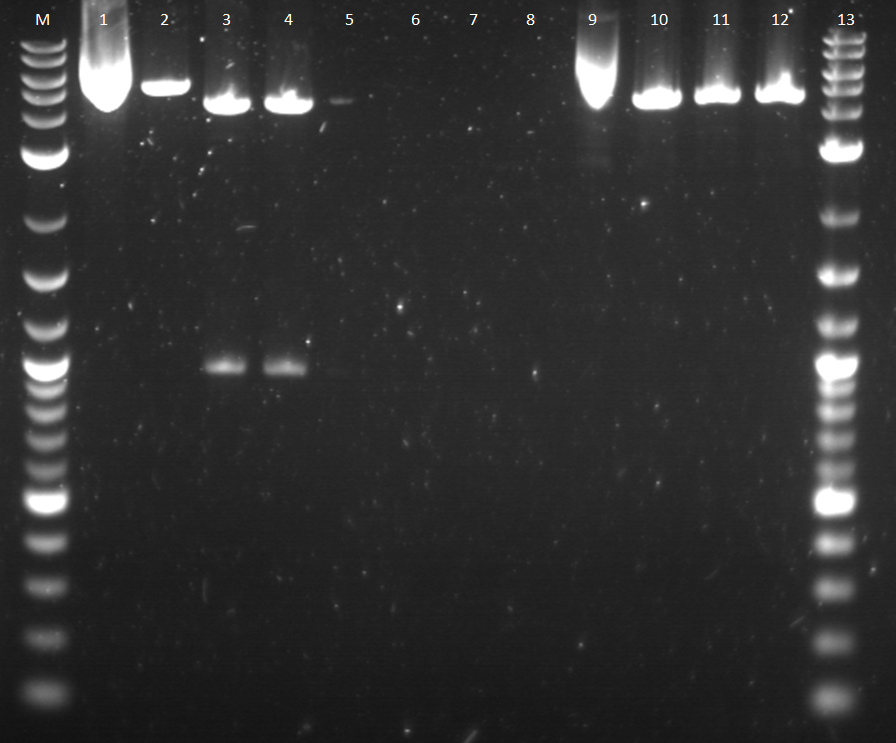
| + | Since the sample construct of pcat_YFPc seemed to not work on the gel or cut correctly. I thought the sample might not have been ligated correctly and contain the restriction site needed. Another restriction digest reaction was made of a double and single cut of the construct, Pcat_YFPc from a different colony. This reaction sample was also ran on a gel. | ||
| + | |||
| + | |||
| + | |||
| + | [[Image: 8_2_11_RD_VY.png | right | 350px |thumb| This is a figure of a gel with multiple cut construct samples.]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | Well 1: BBa_I14033_BBa_E0430 (uncut pcat_YFPc) | ||
| + | Well 2: BBa_I14033_BBa_E0430 (cut pcat_YFPc) | ||
| + | Well 3: BBa_I14033_BBa_E0430 (double cut pcat_YFPc) | ||
| + | |||
| + | Unfortunately, well 3 does not show any bands. Well 2 has a plasmid band that is pretty close to the expect size of the plasmid construct. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | Because the constructs on this plate don't seem to work, I religated pcat with YFPc and transformed it on a new plate. As these reactions were taking place, I also attempted to induce the pbad again. This time various concentrations of arabinose were used on the cell culture of the pbad_YFPc construct, pbad_RFPc construct, and the pbad_GFPc construct. Concentrations of 5 uM, 10 uM, and 50uM. This was for liquid culture in LB and PBS to prevent auto fluorescence from the LB. In theory, it should be easier to see the fluorescence if any in the PBS liquid. | ||
| + | |||
| + | Once there were colonies on the plates for the new pcat_YFPc, plasmid preps were made as well as another restriction digest reaction on them. During restriction digest, I used plasmids from the new plate and old plate. This time a new buffer was also used on the double digestion, Buffer 3 when cutting at X and P sites. Only double cuts were made on these sample with either EcoRI and SpeI sites or XbaI and PstI sites. All these reactions were run on a gel. | ||
| + | |||
| + | |||
| + | [[Image: 8_6_11_RDVY.png | left | 350px |thumb| This is a figure of a gel with multiple cut construct samples.]] | ||
| + | |||
| + | |||
| + | 1. BBa_I14033_BBa_E0430 (uncut) | ||
| + | 2. BBa_I14033_BBa_E0430 (single cut) | ||
| + | 3. BBa_I14033_BBa_E0430 (double cut with EcoRI and SpeI) | ||
| + | 4. BBa_I14033_BBa_E0430 (double cut with XbaI and PstI) | ||
| + | 5. BBa_I14033_BBa_E0430 (uncut) | ||
| + | 6. BBa_I14033_BBa_E0430 (single cut) | ||
| + | 7. BBa_I14033_BBa_E0430 (double cut with EcoRI and SpeI) | ||
| + | 8. BBa_I14033_BBa_E0430 (double cut with XbaI and PstI) | ||
| + | 9. BBa_I14033_BBa_E0430 (uncut) | ||
| + | 10. BBa_I14033_BBa_E0430 (single cut) | ||
| + | 11. BBa_I14033_BBa_E0430 (double cut with EcoRI and SpeI) | ||
| + | 12. BBa_I14033_BBa_E0430 (double cut with XbaI and PstI) | ||
| + | |||
| + | |||
| + | Wells 1-4 were from old colonies while 5-12 were colonies of the new ligation of the same construct. With the new buffer everything seemed to work well and the bands are at the correct sizes for this particular construct. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |} | ||
| + | |||
| + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#333333;margin-top:5px;" cellspacing="18" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h3>Week 10: 8/6-8/12</h3> | ||
| + | |- | ||
| + | | | ||
| + | |||
| + | Since there was success with restriction mapping, I wanted to double check that the construct of which a glycerol stock was made, was correct. Also the insert of the pbad_YFPc plasmid was not checked just the whole size of the plasmid was checked. Therefore, a gel was once again run for all these samples as verification. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | [[Image: 8_9_11_RD_AP_VY.png | right | 350px |thumb| This is a figure of a gel with multiple cut construct samples.]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | 1. BBa_I14033_BBa_E0430 (uncut) | ||
| + | 2. BBa_I14033_BBa_E0430 (single cut) | ||
| + | 3. BBa_I14033_BBa_E0430 (double cut with XbaI and PstI) | ||
| + | 4. BBa_I14033_BBa_E0430 (double cut with EcoRI and SpeI) | ||
| + | 9. BBa_E0430 (double cut with EcoRI and SpeI) | ||
| + | 10.BBa_I3453_BBa_E0430 (double cut with XbaI and PstI) | ||
| + | 11.BBa_I3453_BBa_E0430 (double cut with XbaI and PstI) | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | Because this sample of construct seemed to check out according to restriction mapping, I decided to streak out BBa_I14033_BBa_E0430 from the glycerol stock. The streaked plate was also put under UV light to see if the colonies would glow yellow. They seemed to glow slightly stronger than the original plate of the first ligation; however, it was not as bright as the construct of YFPc with ptet. To verify once again if the streaked plate worked correctly, restriction digest reaction was also set up on the plasmids of the colonies from the streaked plate. The results on the gel seem pretty clear. | ||
| + | |||
| + | |} | ||
| + | |||
| + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#333333;margin-top:5px;" cellspacing="18" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h3>Week 11: 8/13-8/19</h3> | ||
| + | |- | ||
| + | | | ||
| + | |||
| + | My last and final week I decided to experiment with a few other constitutive promoters found in the BBa_J23100 family. The promoters were ligated with the yellow and green fluorescent proteins. The promoters found in J23100 and J23101 seemed to glow the brightest as opposed to the promoter found in J23109. Unfortunately, there was not enough time to collect characterization data to be able to submit the constructs to the parts registry. | ||
| + | |||
| + | This week we also had our final meeting, where we distributed tasks for each team member, to put the final touches to our project. There were several tasks, so there was still a lot of work ahead to do. | ||
| + | |||
| + | |} | ||
Latest revision as of 02:07, 28 September 2011
Vanessa's Notebook
Contents |
Week 1: 6/1-6/10 |
|
The first few days, the group sat through a boot camp, where we learned about biobricks, the basics of Biology such as DNA transcription and translation, and how to use Clotho tools. As exposure to the more computer science aspect of the project, the group learned how to write a Clotho application. As a test run, mini preps (protocol link) were performed on a few samples of biobricks that were previously plated and transformed. Most of these samples unfortunately did not yield a high concentration of DNA, except for a few. After quantifying the samples, the samples were loaded on a gel next to a ladder to confirm the DNA that was present. However, most bands did not appear on the gel after exposed to the UV light. This concluded that there was no DNA in the samples that were being looked at. Next, several transformations were made for biobrick parts such as RBS, RFP, and constituent and inducible promoters. The transformations produced several cultures that were both dark and light in color. Plasmid preps were made of the dark and light cultures of the UV promoter. The dark culture had an okay amount of DNA concentration. However the other samples did not have a good concentration of DNA either. There seemed to be contamination or something else growing on the plates because some of the cultures were oddly shaped and there was a pungent smell. As a result, a sample of plasmid prep culture was stained and looked under the microscope. Most samples did not have E. coli in the sample. |
Week 2: 6/11-6/17 |
|
A second round of plasmid preps were made of the same plates to troubleshoot the cultures that do not have high concentrations of DNA. These unfortunately still did not work or produce anything. Only the UV promoter sample had a culture. This week the BU Wellesley team is demoing the QIAcube from Quiagen. Plasmid prep of the UV sample was used to start up and learn how to use the QIAcube. It seems that the problem is the plates with ampicillin. A sub-group of our team were working on putting together the GFP (Bba_J52028) and terminator (backbone, Bba_B0015) to test out various RBS and promoters combinations. Once the GFP and terminator were ligated together, the construct was transformed on a Kanamycin plate. This successfully grew on the plate. Other parts were transformed and grew nicely on the Kanamycin plates. As a result, it is believed that the ampicilling plates are not selecting because there was also a negative control plated with just top 10 cells and there was some growth seen on the plates. New ampicillin plates were made. New transformations were made of the 7 original parts chosen and few additional ones (RBS, promoters, RFP). Most of these seem to grow successfully, however 2 plates had cultures with a pink tint to them. There was one plate where nothing grew on it. There was also two negative controls for ampicillin plates and kanamycin plates, where nothing grew. Plasmid preps of the new transformations were made.
Wells 1 through 9 and M being the location of the ladder. 1. Bba_I13453 18.6 ng/uL 2. Bba_J23100 53 ng/uL 3. Bba_J61127 23.1 ng/uL 4. Bba_E1010 56.5 ng/uL 5. Bba_J23101 32.6 ng/uL 6. Bba_14053 17.5 ng/uL 7. Bba_J23109 31.6 ng/uL 8. Bba_J61101 9.6 ng/uL 9. Bba_J61100 30.2 ng/uL
|
Week 3: 6/18-6/24 |
|
- Prepared plasmid preps and continued to used the QIAcube for minipreps of biobrick parts listed below:
- Bba_E0430 (YFP composite) 1/A/8I - Bba_E0240 (GFP composite) 1/A/12M - Bba_R2000 (Promoter) 1/K/16F Reporters: - Bba_E0020 (ECFP) 1/A/6A - Bba_E0032 (EYFP) 1/A/6E - Bba_K156010 (BFP) 2/A/4D - Restriction digest on Bba_E0430 (yfpc), Bba_E0240 (gfpc), Bba_R2000 (promoter), Bba_J61127 (RBS), Bba_J61100(RBS), GFP+Term(2) - More plasmid preps of promoter and RBS from 6/15 transformations. - Prepare for joint meeting with Professor Densmore - Gel electrophoresis with restriction digested parts and gel extraction with the QIAcube. Quantification values of the gel extraction were somewhat low. - A restriction digest was performed on BFP and RFP to make up the list of reporter genes that would be ligated with a terminator in route to make one device. - The gel ran once more with digested parts because the bands on the gel looked very faint and did not seem to produce the correct bands according to the parts being cut.
Two approaches were decided on to make the 4 different successful devices, the Bottom Up approach and using composite parts with flourescent proteins to build the some of the devices. The team broke up into two. I became part of the team, where we use the composite parts approach using YFP, GFP, and RFP. I am responsible for making a device with the yellow fluorescent protein composite parts along with four different promoters found in biobrick parts. The promoters include Bba_I13453 (pbad), Bba_I14033 (pcat), Bba_R0040 (ptet), and Bba_R2000.
- Transformation of Bba_R0040 (ptet) - Restriction digest, gel run and extraction with Bba_I13453 (pbad), Bba_I14033 (pcat), and Bba_E0340 (YFPc) - Restriction Digest: Bba_I13453 cut with SpeI and PstI
Five samples were made at 3:1 ratio of insert to backbone.
- pcat + YFPc
- pcat + YFPc
- pbad + YFPc
- pbad + YFPc
- pbad + YFPc
|
Week 4: 6/25-7/1 |
|
1. Restriction digest of Bba_E0430.1 (YFPc) 2. " " " " 3. Uncut Bba_E0430.1 (YFPc) 4. Restriction digest of Bba_E0430.2 (YFPc) 5. " " " " 6. Uncut Bba_E0430.2 (YFPc) 7. Restriction digest of Bba_E0430.2 (YFPc) 8. " " " " 9. Restriction digest of Bba_E0430 (YFPc) 10. " " " " 11. Restriction digest of Bba_R2000
- A two 24 well gel was made with 2 wells per each sample. 10 wells were filled with samples listed above. - These samples were extracted and quantified. The concentration values of the samples were all in single digit. - Ligation: Five samples were ligated at a 6:1 ratio of insert to backbone. - Bba_R2000 + Bba_E0430 - Bba_I14033 + Bba_E0430 - Bba_R0040 + E0430 - Bba_R2000.1.2 + E0430 - Bba_I13453 + E0430
|
Week 5: 7/2-7/8 |
|
Another attempt to transform last week's ligation samples, but unfortunately again no growth. While the ligation issues are being figured out, stocks of parts that are needed for future ligations are being made. Three promoters needed in gel extraction stage are promoters, Bba_I13453, Bba_R0040, and Bba_I14033. These biobrick parts were cut with SpeI and PstI using a higher mass of DNA. These pieces were run on a gel providing some strange unclear results on the image below.
5. Restriction Digest of Bba_I13453.2 6. Uncut Bba_I13453.2 7. Restriction Digest of Bba_R0040 8. Uncut Bba_R0040 9. Restriction Digest of Bba_I14033 10.Uncut Bba_I14033
- Plasmid preps of various parts: Bba_E0430, Bba_I14033, Bba_I13453, Bba_E0240 - Streaked 3 plates from -20 degrees Celsius stock (Bba_R2000, Bba_R0040, and Bba_I13453) |
Week 6: 7/9-7/15 |
|
- Visited MIT - A few of the team members went to talk to MIT's postdoc, Jonathan to troubleshoot our ligation and transformation issues. He suggested checking our gel extraction samples, using a different transformation and ligation protocol. - We thought there may be an issue with out TOP10 competent cells. As a result, we transformed ligation reactions in TOP10 competent cells and commercial cells to see if that was affecting our results. We noticed that once the cells were spun down the pellet of cells was much larger for the commercial cells as opposed to the TOP10 cells made by us. Unfortunately, nothing grew on the plates. - Ligation reactions were attempted again between the YFP composite and various promoters. - Since we thought that the backbones might be re-ligating with itself, we decided to use CIP on the backbone, which dephosphorylates the backbone. These reactions/samples had to be run through a gel, extracted, and quantified. - Ligation reactions were made with CIP on the backbone and without CIP, using quick ligase. These reactions were transformed onto plates, but unfortunately did not grow. |
Week 7: 7/16-7/22 |
|
As a team, we brainstormed ideas on how to find the right combination for ligation protocol between reaction time and ligation ratios. - I tried ligation reactions with Bba_E0430(YFPc) and Bba_R0040(ptet) for 30 minutes, 1 hour, and overnight on the bench. The reactions were the same sample with two different ratios 1:3 and 1:7. I transformed these reactions with TOP10 competent cells. Unfortunately, nothing grew on the plates this try. With the 1:3 ratio, I tried transforming with 4 tubes of TOP10 competent cells, but nothing worked either. - The ligation reaction was transformed with alpha-select competent cells and there was success. A few colonies grew on each plate with various reactions. It grew best at 1:3 for 30 minutes reaction time on bench at room temperature. To confirm these results, 4 plasmid preps were taken from each plate. There were a few with false positives, but the others seem to be at the right position on the gel with okay quantification values. - Two other constructs were ligated and checked on a gel to see if it fits right with what was needed. Unfortunately, the ligation of YFPc and R2000 did not show up on the gel, there must be some kind of error in the reaction. |
Week 8: 7/23-7/29 |
|
Because our ligations seemed to be working successfully, we began to restriction map the various constructs.Before this, the plates with various constructs were checked under UV light. The construct with the constitutive promoter, pcat, BBa_I14033 did not glow under the UV light; however, the construct with ptet_YFPc (BBa_R0040_BBa_E0430) did glow brightly under the UV light. The other constitutive promoter, BBa_R2000 made BBa_E0240 glow green under the UV light. The inducible promoter, pbad (BBa_I13453), had the correct response in which it did not glow because there was no arabinose present. In addition, the constructs were checked by making 4 plasmid preps of each plate, just in case there were false positives on the plate. The plasmids of each plate were then ran on a gel. The constructs tested include:
12-15.BBa_I13453_BBa_E0430 (Pbad_YFPc) 16-19.BBa_I14033_BBa_E0430 (Pcat_YFPc) 20.BBa_R2000_BBa_E0430 (promoter_YFPc)
Also, one of the constructs with promoter pbad is an inducible promoter. Therefore, this construct must be induced with arabinose so it can express the colors, green, red, and in my case yellow. We attempted to induce this promoter by a concentration of arabinose of 50 uM.Unfortunately this did not cause the cell culture to glow.
7-9: BBa_I13453_BBa_E0430 (pbad_YFPc); uncut, cut, double cut (It has the correct size bands) 10-12: BBa_I14033_BBa_E0430 (pcat_YFPc); uncut, cut, double cut (It has incorrect size bands and well 12 does not have two bands. It seems that a double cut was not made.) 13-15: BBa_I14033_BBa_E0240 (pcat_GFPc) uncut, cut, double cut (These bands also seem wrong according to the DNA ladder. The bands for the double cut seem too high)
|
Week 9: 7/30-8/5 |
|
Well 1: BBa_I14033_BBa_E0430 (uncut pcat_YFPc) Well 2: BBa_I14033_BBa_E0430 (cut pcat_YFPc) Well 3: BBa_I14033_BBa_E0430 (double cut pcat_YFPc) Unfortunately, well 3 does not show any bands. Well 2 has a plasmid band that is pretty close to the expect size of the plasmid construct.
Once there were colonies on the plates for the new pcat_YFPc, plasmid preps were made as well as another restriction digest reaction on them. During restriction digest, I used plasmids from the new plate and old plate. This time a new buffer was also used on the double digestion, Buffer 3 when cutting at X and P sites. Only double cuts were made on these sample with either EcoRI and SpeI sites or XbaI and PstI sites. All these reactions were run on a gel.
1. BBa_I14033_BBa_E0430 (uncut) 2. BBa_I14033_BBa_E0430 (single cut) 3. BBa_I14033_BBa_E0430 (double cut with EcoRI and SpeI) 4. BBa_I14033_BBa_E0430 (double cut with XbaI and PstI) 5. BBa_I14033_BBa_E0430 (uncut) 6. BBa_I14033_BBa_E0430 (single cut) 7. BBa_I14033_BBa_E0430 (double cut with EcoRI and SpeI) 8. BBa_I14033_BBa_E0430 (double cut with XbaI and PstI) 9. BBa_I14033_BBa_E0430 (uncut) 10. BBa_I14033_BBa_E0430 (single cut) 11. BBa_I14033_BBa_E0430 (double cut with EcoRI and SpeI) 12. BBa_I14033_BBa_E0430 (double cut with XbaI and PstI)
|
Week 10: 8/6-8/12 |
|
Since there was success with restriction mapping, I wanted to double check that the construct of which a glycerol stock was made, was correct. Also the insert of the pbad_YFPc plasmid was not checked just the whole size of the plasmid was checked. Therefore, a gel was once again run for all these samples as verification.
1. BBa_I14033_BBa_E0430 (uncut) 2. BBa_I14033_BBa_E0430 (single cut) 3. BBa_I14033_BBa_E0430 (double cut with XbaI and PstI) 4. BBa_I14033_BBa_E0430 (double cut with EcoRI and SpeI) 9. BBa_E0430 (double cut with EcoRI and SpeI) 10.BBa_I3453_BBa_E0430 (double cut with XbaI and PstI) 11.BBa_I3453_BBa_E0430 (double cut with XbaI and PstI)
|
Week 11: 8/13-8/19 |
|
My last and final week I decided to experiment with a few other constitutive promoters found in the BBa_J23100 family. The promoters were ligated with the yellow and green fluorescent proteins. The promoters found in J23100 and J23101 seemed to glow the brightest as opposed to the promoter found in J23109. Unfortunately, there was not enough time to collect characterization data to be able to submit the constructs to the parts registry. This week we also had our final meeting, where we distributed tasks for each team member, to put the final touches to our project. There were several tasks, so there was still a lot of work ahead to do. |
 "
"