Team:ENSPS-Strasbourg/Background
From 2011.igem.org
(→Abstraction level) |
|||
| (11 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{ENSPS-Strasbourg/Temp}} | {{ENSPS-Strasbourg/Temp}} | ||
| + | __NOTOC__ | ||
| + | <span id="proj"></span> | ||
=Introduction= | =Introduction= | ||
<DIV STYLE="text-align:justify;font-weight:bold;"> | <DIV STYLE="text-align:justify;font-weight:bold;"> | ||
| Line 6: | Line 8: | ||
</DIV> | </DIV> | ||
| - | = | + | |
| - | + | =Global project:= | |
| - | + | The final aim of our project is to create a complete automated design tool to help the biologist during the whole Biosystem’s design flow : | |
| - | + | ||
| - | In this way, our | + | [[File:detailled_scheme.png|border|800px|center|link=]] |
| - | </ | + | |
| + | The above scheme represents the final software suite we want to develop. Everything starts with an idea of a Biosystem, or a biological function, which leads to write some specifications and requirements. Then, as the microelectronics design flow uses HDL (High-level Description Languages), we could imagine that a description of this biosystem will be written with a “biologic description language” (best language to be defined). Then, a tool will translate it into a block diagram of elementary functions, in a high description level (only with input and output species, and different biological mechanisms). Another tool will find the best suitable BioBricks to realize the system, linked with a BioBrick registration database, which will give to the biologist the BioBricks involved into the system. | ||
| + | Another part of this suite is to simulate the biosystem before building it. For that, and in using analogies between electronics and biology, the biosystem will be translated into an electronical equivalent, and this equivalent will be simulated into a conventional electronic simulator. Here, two simulations will be possible. Firstly, a behavioral simulation (binary model) will give to the biologist an idea of the behavior of the system, in putting different parameters (species present or not at the beginning for example). This simulation will validate the proof of concept. After that, another simulation will simulates in continuous-time the system (conservative model), and here the biologist will see the different interaction between species, the evolution of the concentrations into the system, and have an accurate idea of the transient states.<span id="first"></span> | ||
| + | This view of our work is an ideal view, and we are only at the beginning of the elaboration of this software tool. For us, this year, the aim of our work is to prove that a biological system can be simulated with an electronic equivalent by a conventional electronic simulator. | ||
| + | |||
| + | |||
| + | =Our first step:= | ||
| + | The initial part of our project is to prove that electronics and biology could be merged so as to build this kind of software tool. This year, we have decided to be focused on a crucial part of our project which is the simulation of biological mechanisms thanks to electronic tools. Then, by proving analogies between electronic and biological systems, we have built a software which simulates a system of biological devices as an electronic system. | ||
| + | In other words, our work was aimed at translating biological mechanisms into a specific language understandable by conventional electronic simulators, so as to be able to virtually build a biosystem, and simulate it. In this way, we can take advantage of the powerfulness of electronic simulators for simulating complex systems in a reduced time. | ||
| + | The global scheme of our year’s work is available below: | ||
| + | |||
| + | [[File:this_year_scheme.png|border|600px|center|link=]] | ||
| + | |||
| + | As we can see above, the first step is to make different analogies between electronics and biology to have electronic models of biological mechanisms (all is visible in the Modeling part). The idea is to develop a software in which the biologist enters the different species involved, and adds different mechanisms. Here, the level of abstraction is raised, as seen in the next part. Then, the system is translated and simulated as an electronic device. To have a better idea of how the software is build, you can go to the Software part. | ||
| + | |||
| + | ==Abstraction level== | ||
| + | To have an efficient tool, and to be as general as possible, we have decided to increase the abstraction level for designing the system. Indeed, today the research works are always made at a “BioBrick part” level of abstraction. Then more and more BioBricks are created, and some of them realize the same function. In parallel, the most efficiently characterized BioBricks could be reproduced, and then biologists can increase the number of “biobrick parts” to build reliable “biodevices”. But while the complexity of those “biodevices” is increasing, it appears more and more difficult to hand-make a complex scheme, with all the species involved into the system, and to pay attention to all the interactions possible between these species. Moreover, to physically build a BioBrick is time and money consuming.<br> | ||
| + | |||
| + | Then, if we increase the level of abstraction of the “BioSystem creation”, in forgetting the BioBrick parts, and in focusing on the mechanisms and the different species involved, our tool enables a biologist to create and simulate his system, and to have an accurate idea of the behavior of it.<br> | ||
| + | |||
| + | We have so decided to talk only about three basic mechanisms : <b>Inhibition</b> - <b>Association</b> (or Complex reaction) - and <b>Syhthesis</b> | ||
Latest revision as of 18:47, 28 October 2011
Introduction
Imagine that you could build a complete biosystem, with many biobricks involved in it, without complex laboratories experiences, fastidious studies, and in taking advantage of a complete Automation Design Software suite.
Global project:
The final aim of our project is to create a complete automated design tool to help the biologist during the whole Biosystem’s design flow :
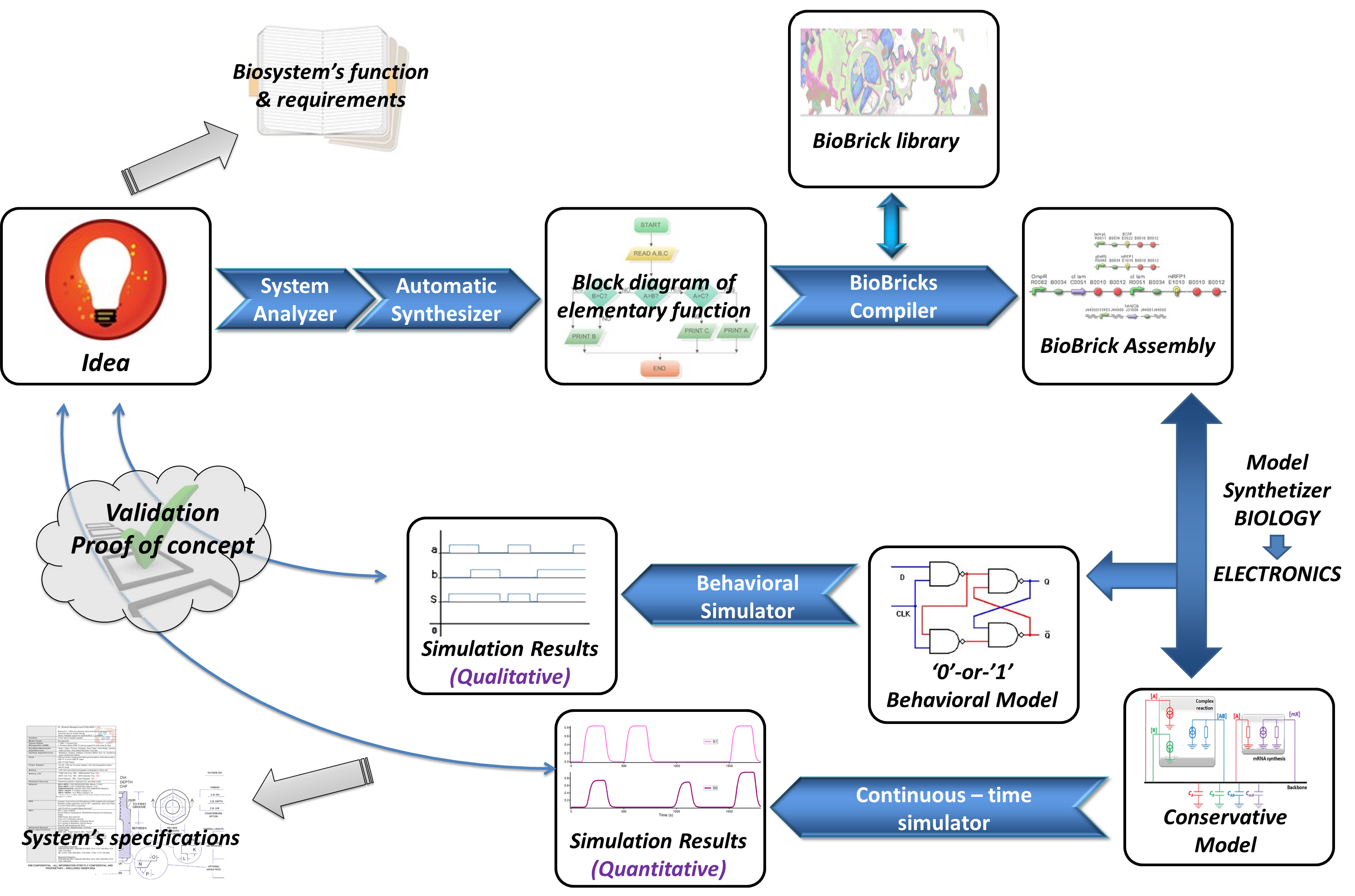
The above scheme represents the final software suite we want to develop. Everything starts with an idea of a Biosystem, or a biological function, which leads to write some specifications and requirements. Then, as the microelectronics design flow uses HDL (High-level Description Languages), we could imagine that a description of this biosystem will be written with a “biologic description language” (best language to be defined). Then, a tool will translate it into a block diagram of elementary functions, in a high description level (only with input and output species, and different biological mechanisms). Another tool will find the best suitable BioBricks to realize the system, linked with a BioBrick registration database, which will give to the biologist the BioBricks involved into the system. Another part of this suite is to simulate the biosystem before building it. For that, and in using analogies between electronics and biology, the biosystem will be translated into an electronical equivalent, and this equivalent will be simulated into a conventional electronic simulator. Here, two simulations will be possible. Firstly, a behavioral simulation (binary model) will give to the biologist an idea of the behavior of the system, in putting different parameters (species present or not at the beginning for example). This simulation will validate the proof of concept. After that, another simulation will simulates in continuous-time the system (conservative model), and here the biologist will see the different interaction between species, the evolution of the concentrations into the system, and have an accurate idea of the transient states. This view of our work is an ideal view, and we are only at the beginning of the elaboration of this software tool. For us, this year, the aim of our work is to prove that a biological system can be simulated with an electronic equivalent by a conventional electronic simulator.
Our first step:
The initial part of our project is to prove that electronics and biology could be merged so as to build this kind of software tool. This year, we have decided to be focused on a crucial part of our project which is the simulation of biological mechanisms thanks to electronic tools. Then, by proving analogies between electronic and biological systems, we have built a software which simulates a system of biological devices as an electronic system. In other words, our work was aimed at translating biological mechanisms into a specific language understandable by conventional electronic simulators, so as to be able to virtually build a biosystem, and simulate it. In this way, we can take advantage of the powerfulness of electronic simulators for simulating complex systems in a reduced time. The global scheme of our year’s work is available below:
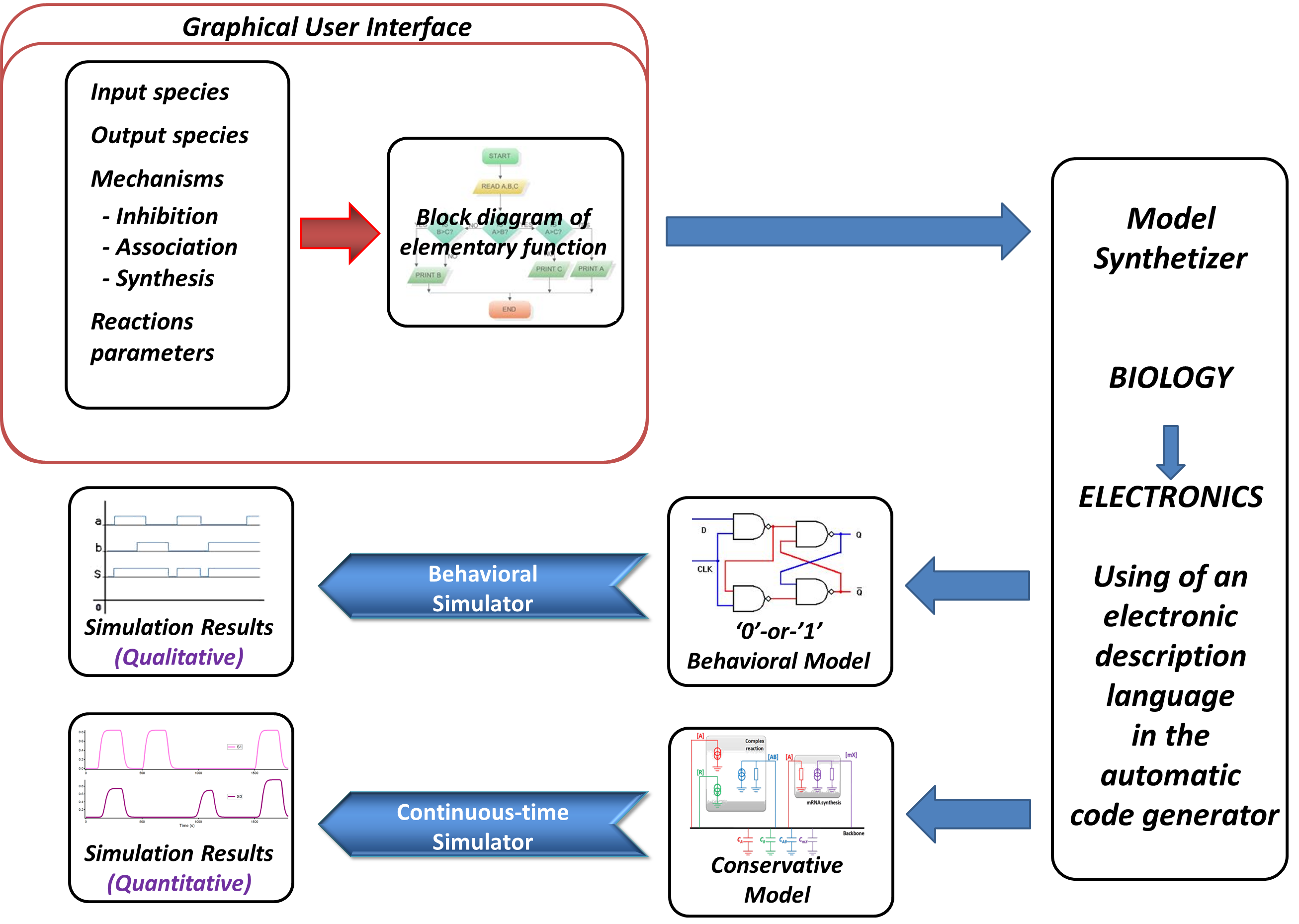
As we can see above, the first step is to make different analogies between electronics and biology to have electronic models of biological mechanisms (all is visible in the Modeling part). The idea is to develop a software in which the biologist enters the different species involved, and adds different mechanisms. Here, the level of abstraction is raised, as seen in the next part. Then, the system is translated and simulated as an electronic device. To have a better idea of how the software is build, you can go to the Software part.
Abstraction level
To have an efficient tool, and to be as general as possible, we have decided to increase the abstraction level for designing the system. Indeed, today the research works are always made at a “BioBrick part” level of abstraction. Then more and more BioBricks are created, and some of them realize the same function. In parallel, the most efficiently characterized BioBricks could be reproduced, and then biologists can increase the number of “biobrick parts” to build reliable “biodevices”. But while the complexity of those “biodevices” is increasing, it appears more and more difficult to hand-make a complex scheme, with all the species involved into the system, and to pay attention to all the interactions possible between these species. Moreover, to physically build a BioBrick is time and money consuming.
Then, if we increase the level of abstraction of the “BioSystem creation”, in forgetting the BioBrick parts, and in focusing on the mechanisms and the different species involved, our tool enables a biologist to create and simulate his system, and to have an accurate idea of the behavior of it.
We have so decided to talk only about three basic mechanisms : Inhibition - Association (or Complex reaction) - and Syhthesis
 "
"