Team:USTC-China/Project/module
From 2011.igem.org
(→Quorum Sensing) |
(→Quorum Sensing) |
||
| (12 intermediate revisions not shown) | |||
| Line 11: | Line 11: | ||
==Riboswitch== | ==Riboswitch== | ||
<html> | <html> | ||
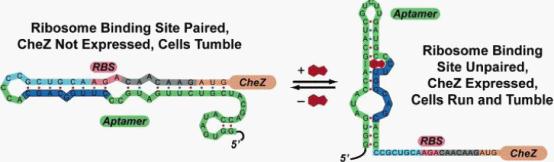
| - | <div class="center"><div class="thumb tnone"><div class="thumbinner" ><img alt="Riboswitch.jpg" src="/wiki/images/3/36/Riboswitch.jpg" width="554" height="162" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Riboswitch.jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" alt="" /></a></div></div></div></div></div><br /> | + | <div class="center"><div class="thumb tnone"><div class="thumbinner" ><img alt="Riboswitch.jpg" src="/wiki/images/3/36/Riboswitch.jpg" width="554" height="162" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Riboswitch.jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" alt="" /></a></div>Figure 1: Model for how the theophylline-sensitive synthetic riboswitch controls the translation of the CheZ protein.</div></div></div></div><br /> |
</html> | </html> | ||
| - | |||
| - | |||
In the absence of theophylline (left),the mRNA adopts a conformation in which the ribosome binding site is paired and translation of CheZ is inhibited. In the absence of CheZ, the protein CheY-P remains phosphorylated and the cells tumble in place. In the presence of theophylline (shown in red), the mRNA can adopt a conformation in which the ribosome binding site is exposed and CheZ is expressed, thus allowing the cells to run and tumble. (Images from: Shana Topp, Justin P. Gallivan (2006) Guiding Bacteria with Small Molecules and RNA. J.Am.Chem.Soc, 129,6870-6811) | In the absence of theophylline (left),the mRNA adopts a conformation in which the ribosome binding site is paired and translation of CheZ is inhibited. In the absence of CheZ, the protein CheY-P remains phosphorylated and the cells tumble in place. In the presence of theophylline (shown in red), the mRNA can adopt a conformation in which the ribosome binding site is exposed and CheZ is expressed, thus allowing the cells to run and tumble. (Images from: Shana Topp, Justin P. Gallivan (2006) Guiding Bacteria with Small Molecules and RNA. J.Am.Chem.Soc, 129,6870-6811) | ||
| + | |||
| + | <br/> | ||
<html><a name="Toggle-switch" id=""></a></html> | <html><a name="Toggle-switch" id=""></a></html> | ||
==Toggle-switch== | ==Toggle-switch== | ||
<html> | <html> | ||
| - | <div class="center" ><div class="thumb tnone" style="width:554px;"><div class="thumbinner" style="width:554px;"><a href="/File:Toggle-switch.jpg" class="image"><img alt="Toggle-switch.jpg" src="/wiki/images/7/79/Toggle-switch.jpg" width="554" height="62"></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Toggle-switch.jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div></div></div></div></div> | + | <div class="center" ><div class="thumb tnone" style="width:554px;"><div class="thumbinner" style="width:554px;"><a href="/File:Toggle-switch.jpg" class="image"><img alt="Toggle-switch.jpg" src="/wiki/images/7/79/Toggle-switch.jpg" width="554" height="62"></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Toggle-switch.jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure 2: Toggle switches incorporate three mechanisms: positive feedback, double-negative feedback, and the repressor binding cooperatively and two different kinds of bacteria differentiate out of homogeneous conditions. (Images from: Chunbo Lou, Xili Liu, Ming Ni.etc (2010) Synthesizing a novel genetic sequential logic circuit: a push-on push-off switch. Molecular Systems Biology, 6:350)</div></div></div></div> |
| + | |||
| - | |||
</html> | </html> | ||
<br/> | <br/> | ||
<html> | <html> | ||
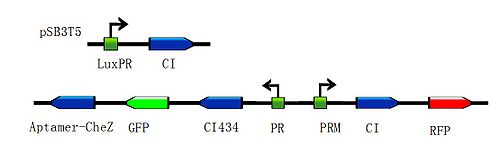
| - | <div class="center" ><div class="thumb tnone" style="width:502px;"><div class="thumbinner" style="width:502px;"><a href="/File:Toggle-switch1.jpg" class="image"><img alt="Toggle-switch1.jpg" src="/wiki/images/thumb/a/ab/Toggle-switch1.jpg/500px-Toggle-switch1.jpg" width="500" height="155" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Toggle-switch1.jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div></div></div></div></div> | + | <div class="center" ><div class="thumb tnone" style="width:502px;"><div class="thumbinner" style="width:502px;"><a href="/File:Toggle-switch1.jpg" class="image"><img alt="Toggle-switch1.jpg" src="/wiki/images/thumb/a/ab/Toggle-switch1.jpg/500px-Toggle-switch1.jpg" width="500" height="155" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Toggle-switch1.jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure 3: Constructions to modulate the ratio of toggle switches. The above bar represents pSB3T5, a low to medium copy BioBrick standard vector, where LuxpR (BBa_R0062) controls the expression of cI repressor which would influence the ratio of toggle switch. (Images by USTC-IGEM with TinkerCell)</div></div></div></div> |
</html> | </html> | ||
| - | + | ||
| Line 37: | Line 37: | ||
==Quorum Sensing== | ==Quorum Sensing== | ||
<html> | <html> | ||
| - | <div class="center"><div class="thumb tnone" style="width:554px;"><div class="thumbinner" style="width:554px;"><a href="/File:Quorum_sensing.jpg" class="image"><img alt="Quorum sensing.jpg" src="/wiki/images/5/5c/Quorum_sensing.jpg" width="554" height="269" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Quorum_sensing.jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div></div></div></div></div> | + | <div class="center"><div class="thumb tnone" style="width:554px;"><div class="thumbinner" style="width:554px;"><a href="/File:Quorum_sensing.jpg" class="image"><img alt="Quorum sensing.jpg" src="/wiki/images/5/5c/Quorum_sensing.jpg" width="554" height="269" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Quorum_sensing.jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure 4: The quorum-sensing paradigm in Gram-negative bacteria.</div></div></div></div> |
</html> | </html> | ||
| - | + | a. At low bacterial cell densities AHL molecules are synthesized and accumulate. Depending on the length of the acyl chain, AHLs either diffuse or are pumped out of the cell into the local environment, where the molecules are available for diffusion into, or uptake by, bacterial cells. b. At high bacterial cell densities, a threshold concentration of AHL molecules has accumulated. The R protein forms a complex with its cognate AHL and this complex either activates or represses the transcription of target genes. As the I gene is usually positively regulated by the R protein in complex with the cognate AHL, rapid amplification of the AHL signal results, which facilitates the coordinate transcriptional regulation of multiple genes. (Images from: Andrée M. Lazdunski, Isabelle Ventre and James N. Sturgis(2004) Regulatory circuits and communication in Gram-negative bacteria. Microbiology, Volume 2, July 2004, 581-592) | |
<html> | <html> | ||
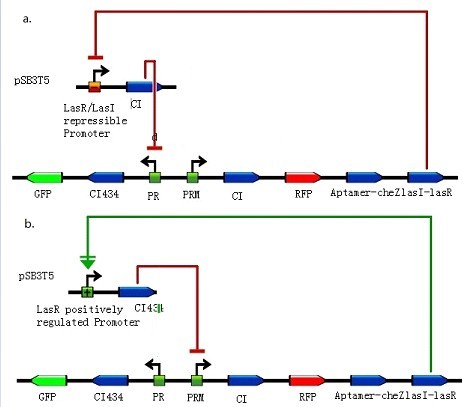
| - | <div class="center"><div class="thumb tnone" style="width:463px;"><div class="thumbinner" style="width:463px;"><a href="/File:Quorum_sensing1.jpg" class="image"><img alt="Quorum sensing1.jpg" src="/wiki/images/2/2d/Quorum_sensing1.jpg" width="463" height="407" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Quorum_sensing1.jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div></div></div></div></div> | + | <div class="center"><div class="thumb tnone" style="width:463px;"><div class="thumbinner" style="width:463px;"><a href="/File:Quorum_sensing1.jpg" class="image"><img alt="Quorum sensing1.jpg" src="/wiki/images/2/2d/Quorum_sensing1.jpg" width="463" height="407" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Quorum_sensing1.jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure 5: Integration of quorum sensing to help grouping.</div></div></div></div> |
</html> | </html> | ||
| - | + | a. LasR/LasI repressible promoter and cI assembled on pSB3T5. In order to increase the ratio of bacteria moving away, we insert aptamer-CheZ into the other side of toggle switch compared with experiments earlier. With lasI-lasR accumulating, LasR/LasI repressible promoter in bacteria which express CheZ and move along theophylline gradient would get repressed and less cI might push off toggle switch to the other state. b. LasR positively regulated promoter and cI434 assembled on pSB3T5. Another design concerns with a LasR positively regulated promoter, which would be activated and express cI434 in bacteria expressing CheZ and also push off toggle switch to the other state. (Images by USTC-IGEM with TinkerCell) | |
<html><a name="Artificial" id=""></a></html> | <html><a name="Artificial" id=""></a></html> | ||
| - | ==Artificial | + | ==Artificial Bacterial Barrier== |
<html> | <html> | ||
| - | <div class="center"><div class="thumb tnone" style="width:389px;"><div class="thumbinner" style="width:389px;"><a href="/File:V().jpg" class="image"><img alt="350" src="/wiki/images/a/a6/V%28%29.jpg" width="389" height="248" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:V().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div></div></div></div></div> | + | <div class="center"><div class="thumb tnone" style="width:389px;"><div class="thumbinner" style="width:389px;"><a href="/File:V().jpg" class="image"><img alt="350" src="/wiki/images/a/a6/V%28%29.jpg" width="389" height="248" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:V().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure7:By using the quorum sensing, the engineered bacteria can express Pyosin and Lysis proteins in the high AHL concentration condition, then these bacteria will destroy themselves and release the Pyosin to kill the pathogens(Image from Nazanin Saeidi, Choon Kit Wong, Tat-Ming Lo .etc (2011)).</div></div></div></div> |
</html> | </html> | ||
| - | <p> | + | <p></p> |
<html> | <html> | ||
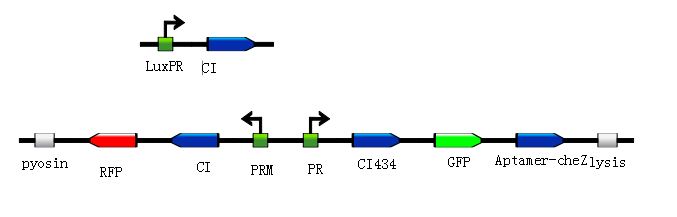
| - | <div class="center"><div class="thumb tnone" style="width:677px;"><div class="thumbinner" style="width:677px;"><a href="/File:J().jpg" class="image"><img alt="250" src="/wiki/images/f/f8/J%28%29.jpg" width="677" height="217" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:J().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div></div></div></div></div> | + | <div class="center"><div class="thumb tnone" style="width:677px;"><div class="thumbinner" style="width:677px;"><a href="/File:J().jpg" class="image"><img alt="250" src="/wiki/images/f/f8/J%28%29.jpg" width="677" height="217" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:J().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure8.Simulated experiment construction</div></div></div></div> |
</html> | </html> | ||
| - | <p> | + | <p>According to the basic design and we assume the small molecule is theophylline, so under the control of this module, the reprogrammed bacteria with green fluorescence will move toward the infection site with a high concentration of theophylline and the expression of Lysis becomes more and more, while the reprogrammed bacteria with red fluorescence will stay at the original site and wait to creat the second wave of the reprogrammed bacteria with green fluorescence, they have been protected from being destroyed by the pyosin from the reprogrammed bacteria with green fluorescence. The reprogrammed bacteria with green fluorescence will destroy themselves and release the Pyosin, which is expressed at the beginning of the process, to kill the pathogens.(Images by USTC-IGEM with TinkerCell)</p> |
Latest revision as of 23:11, 28 October 2011

Contents |
Modules
1.Riboswitch
2.Toggle-switch
3.Quorum Sensing
4.Artificial Innate Immunity System
Riboswitch
In the absence of theophylline (left),the mRNA adopts a conformation in which the ribosome binding site is paired and translation of CheZ is inhibited. In the absence of CheZ, the protein CheY-P remains phosphorylated and the cells tumble in place. In the presence of theophylline (shown in red), the mRNA can adopt a conformation in which the ribosome binding site is exposed and CheZ is expressed, thus allowing the cells to run and tumble. (Images from: Shana Topp, Justin P. Gallivan (2006) Guiding Bacteria with Small Molecules and RNA. J.Am.Chem.Soc, 129,6870-6811)
Toggle-switch

Quorum Sensing
a. At low bacterial cell densities AHL molecules are synthesized and accumulate. Depending on the length of the acyl chain, AHLs either diffuse or are pumped out of the cell into the local environment, where the molecules are available for diffusion into, or uptake by, bacterial cells. b. At high bacterial cell densities, a threshold concentration of AHL molecules has accumulated. The R protein forms a complex with its cognate AHL and this complex either activates or represses the transcription of target genes. As the I gene is usually positively regulated by the R protein in complex with the cognate AHL, rapid amplification of the AHL signal results, which facilitates the coordinate transcriptional regulation of multiple genes. (Images from: Andrée M. Lazdunski, Isabelle Ventre and James N. Sturgis(2004) Regulatory circuits and communication in Gram-negative bacteria. Microbiology, Volume 2, July 2004, 581-592)
a. LasR/LasI repressible promoter and cI assembled on pSB3T5. In order to increase the ratio of bacteria moving away, we insert aptamer-CheZ into the other side of toggle switch compared with experiments earlier. With lasI-lasR accumulating, LasR/LasI repressible promoter in bacteria which express CheZ and move along theophylline gradient would get repressed and less cI might push off toggle switch to the other state. b. LasR positively regulated promoter and cI434 assembled on pSB3T5. Another design concerns with a LasR positively regulated promoter, which would be activated and express cI434 in bacteria expressing CheZ and also push off toggle switch to the other state. (Images by USTC-IGEM with TinkerCell)
Artificial Bacterial Barrier
According to the basic design and we assume the small molecule is theophylline, so under the control of this module, the reprogrammed bacteria with green fluorescence will move toward the infection site with a high concentration of theophylline and the expression of Lysis becomes more and more, while the reprogrammed bacteria with red fluorescence will stay at the original site and wait to creat the second wave of the reprogrammed bacteria with green fluorescence, they have been protected from being destroyed by the pyosin from the reprogrammed bacteria with green fluorescence. The reprogrammed bacteria with green fluorescence will destroy themselves and release the Pyosin, which is expressed at the beginning of the process, to kill the pathogens.(Images by USTC-IGEM with TinkerCell)
 "
"