Team:XMU-China/Project/Description
From 2011.igem.org
(Difference between revisions)
(→Project Description) |
|||
| (3 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
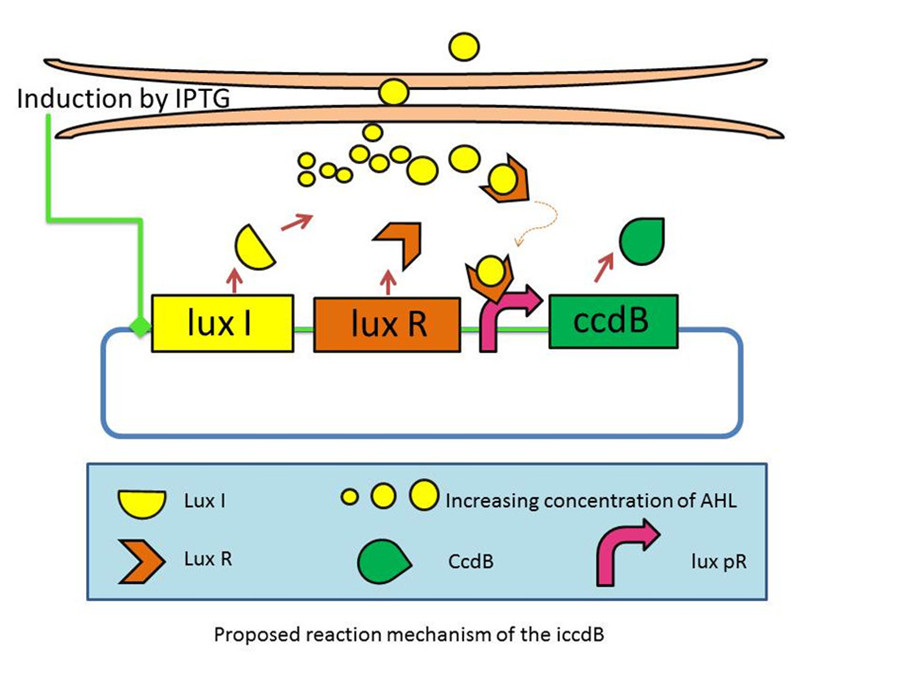
| - | We have developed a series of devices which program a bacteria population to maintain at different cell densities. We have designed and characterized the genetic circuit to establish a bacterial ‘population-control’ device in ''E. coli'' based on the well-known quorum-sensing system from ''Vibrio fischeri'', which autonomously regulates the density of an ''E. coli'' population. The cell density however is influenced by the expression levels of a killer gene (''ccdB'') in our device. As such, we have successfully controlled the expression levels of ''ccdB'' by | + | We have developed a series of devices which program a bacteria population to maintain at different cell densities. We have designed and characterized the genetic circuit to establish a bacterial ‘population-control’ device in ''E. coli'' based on the well-known quorum-sensing system from ''Vibrio fischeri'', which autonomously regulates the density of an ''E. coli'' population. The cell density however is influenced by the expression levels of a killer gene (''ccdB'') in our device. As such, we have successfully controlled the expression levels of ''ccdB'' by using RBSes of different strength and mutated ''luxR'' promoters (''lux pr''). We are working on builting up a database for a series of mutation sites and RBSes corresponding to different steady-state cell densities. An artificial neural network will be built to model and predict the cell density of an ''E. coli'' population. This work can serve as a foundation for future advances involving fermentation industry and information processing. |
[[Image:XMU_China_133beta2.jpg|left|950px|Figure 1 The bacteria population-control device|frame|Figure 1 The bacteria population-control device.]] | [[Image:XMU_China_133beta2.jpg|left|950px|Figure 1 The bacteria population-control device|frame|Figure 1 The bacteria population-control device.]] | ||
| + | |||
| + | <html> | ||
| + | <img src="http://partsregistry.org/wiki/images/4/41/XMU_China_block.jpg"> | ||
| + | </html> | ||
| + | |||
This circuit incorporates a mechanism for programmed death in respond to changes in the environment, and allows us to probe the design principles of its more complex natural counterparts. | This circuit incorporates a mechanism for programmed death in respond to changes in the environment, and allows us to probe the design principles of its more complex natural counterparts. | ||
| + | |||
| + | ==reference== | ||
| + | [1] Kempner E, Hanson F. Aspects of light production by Photobacterium fischeri[J]. Journal of Bacteriology, 1968, 95(3): 975-979. | ||
| + | |||
| + | [2] Fuqua WC, Winan SC, Greenberg E. Quorum sensing in bacteria: the luxR-luxI family of cell density-responsive transcriptional regulators[J]. Journal of Bacteriology, 1994, 176(2): 269-275. | ||
| + | |||
| + | [3] Baldwin T, Devine JH, Heckel, RC, Lin, JW, Shadel GS. The complete nucleotide sequence of the lux regulon of Vibrio fischeri and the luxABN region of Photobacterium leiognathi and the mechanism of control of bacterial bioluminescence[J]. Journal of Bioluminescence and Chemiluminescence, 1989, 4(1): 326-341. | ||
| + | |||
| + | [4] You L, Cox RS, Weiss R, Arnold FH. Programmed population control by cell-cell communication and regulated killing[J]. Nature, 2004, 428(6985): 868-871. | ||
| + | |||
| + | [5] Kampranis SC, Howells AJ, Maxwell A. The interaction of DNA gyrase with the bacterialtoxin CcdB: evidence for the existence of two gyrase-CcdB complexes[J]. Journal of Molecular Biology, 1999, 293(3): 733-744. | ||
Latest revision as of 03:48, 29 October 2011
 "
"