Team:Peking S/project
From 2011.igem.org
| (9 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
{{https://2011.igem.org/Team:Peking_S/back2}} | {{https://2011.igem.org/Team:Peking_S/back2}} | ||
{{Template:Http://2011.igem.org/Team:Peking_S/box4project}} | {{Template:Http://2011.igem.org/Team:Peking_S/box4project}} | ||
| - | |||
| Line 9: | Line 8: | ||
<font color="#ffffff"> <font size=6>Project Description</font></font> | <font color="#ffffff"> <font size=6>Project Description</font></font> | ||
<br><br><br> | <br><br><br> | ||
| - | |||
| - | |||
'''Cell-cell communication-based multicellular networks provide an extended vista for synthetic biology'''. By compartmentalizing complex genetic circuits into separate engineered cells, the difficulty of the construction by layering elementary gates can be dramatically reduced, partly due to the insulation of crosstalk between modules, the suppression of noise by populationally averaging, and the reducing of metabolic burden in host cells. What’s more, cell-cell communication-based multicellular feature enables coordination and synchronization among cells in and between populations and facilitates the generation of reliable non-Boolean dynamics. | '''Cell-cell communication-based multicellular networks provide an extended vista for synthetic biology'''. By compartmentalizing complex genetic circuits into separate engineered cells, the difficulty of the construction by layering elementary gates can be dramatically reduced, partly due to the insulation of crosstalk between modules, the suppression of noise by populationally averaging, and the reducing of metabolic burden in host cells. What’s more, cell-cell communication-based multicellular feature enables coordination and synchronization among cells in and between populations and facilitates the generation of reliable non-Boolean dynamics. | ||
| - | |||
| + | <center>[[File:PkuXor1.png|340px]]</center> | ||
| - | |||
| + | However, orthogonal ‘chemical wires’ that allow concurrent communication are far from sufficient. Accordingly, '''our project intends to develop a versatile ‘chemical wire’ toolbox for both multicellular Boolean computing and non-Boolean dynamics.''' Criteria and assays for ‘chemical wire’ characterization and validation have been established, in which the orthogonality, the speed of signal relaying and the ability to coordinate cells are verified. A set of recently reported quorum sensing systems have been selected, together with previously investigated AHL system, as ‘chemical wire’ toolbox. With this toolbox, multicellular design principles for complex logic circuit that allow a reduction in the complexity and a guarantee in both robust function and operation speed of a system will be proposed, followed by constructing reusable E.coli cell types that each compartmentalizes specific elementary logic gate in a single cell, connected by ‘chemical wires’. By layering a very few number of such cellular logic gates, complex logic computing process will be easily implemented. Besides, a hierarchical population density balancer was constructed, in order to verify the feasibility of our ‘chemical wire’ toolkit for non-Boolean dynamic functions. Nonetheless, challenges remain for this non-Boolean process because communication signals are produced by growing cells, i.e. changes in cell densities may affect the strength of communication. To crack these hurdles, we will develop a microfluidic device that imposes constraints on cell densities and on the flow of chemical signals to guarantee well-coordinated population dynamics. | ||
| + | [[File:Pkusdes1.png|620px]] | ||
<html> | <html> | ||
Latest revision as of 03:50, 6 October 2011
Template:Https://2011.igem.org/Team:Peking S/bannerhidden Template:Https://2011.igem.org/Team:Peking S/back2
Template:Https://2011.igem.org/Team:Peking S/bannerhidden

Project Description
Cell-cell communication-based multicellular networks provide an extended vista for synthetic biology. By compartmentalizing complex genetic circuits into separate engineered cells, the difficulty of the construction by layering elementary gates can be dramatically reduced, partly due to the insulation of crosstalk between modules, the suppression of noise by populationally averaging, and the reducing of metabolic burden in host cells. What’s more, cell-cell communication-based multicellular feature enables coordination and synchronization among cells in and between populations and facilitates the generation of reliable non-Boolean dynamics.
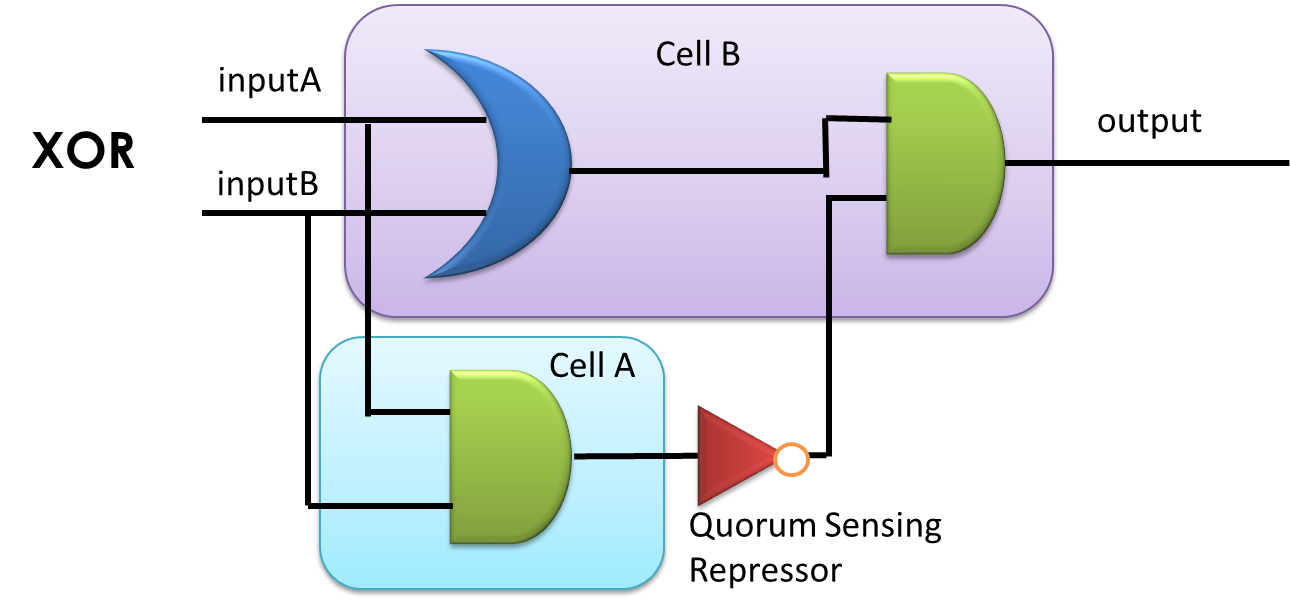
However, orthogonal ‘chemical wires’ that allow concurrent communication are far from sufficient. Accordingly, our project intends to develop a versatile ‘chemical wire’ toolbox for both multicellular Boolean computing and non-Boolean dynamics. Criteria and assays for ‘chemical wire’ characterization and validation have been established, in which the orthogonality, the speed of signal relaying and the ability to coordinate cells are verified. A set of recently reported quorum sensing systems have been selected, together with previously investigated AHL system, as ‘chemical wire’ toolbox. With this toolbox, multicellular design principles for complex logic circuit that allow a reduction in the complexity and a guarantee in both robust function and operation speed of a system will be proposed, followed by constructing reusable E.coli cell types that each compartmentalizes specific elementary logic gate in a single cell, connected by ‘chemical wires’. By layering a very few number of such cellular logic gates, complex logic computing process will be easily implemented. Besides, a hierarchical population density balancer was constructed, in order to verify the feasibility of our ‘chemical wire’ toolkit for non-Boolean dynamic functions. Nonetheless, challenges remain for this non-Boolean process because communication signals are produced by growing cells, i.e. changes in cell densities may affect the strength of communication. To crack these hurdles, we will develop a microfluidic device that imposes constraints on cell densities and on the flow of chemical signals to guarantee well-coordinated population dynamics.
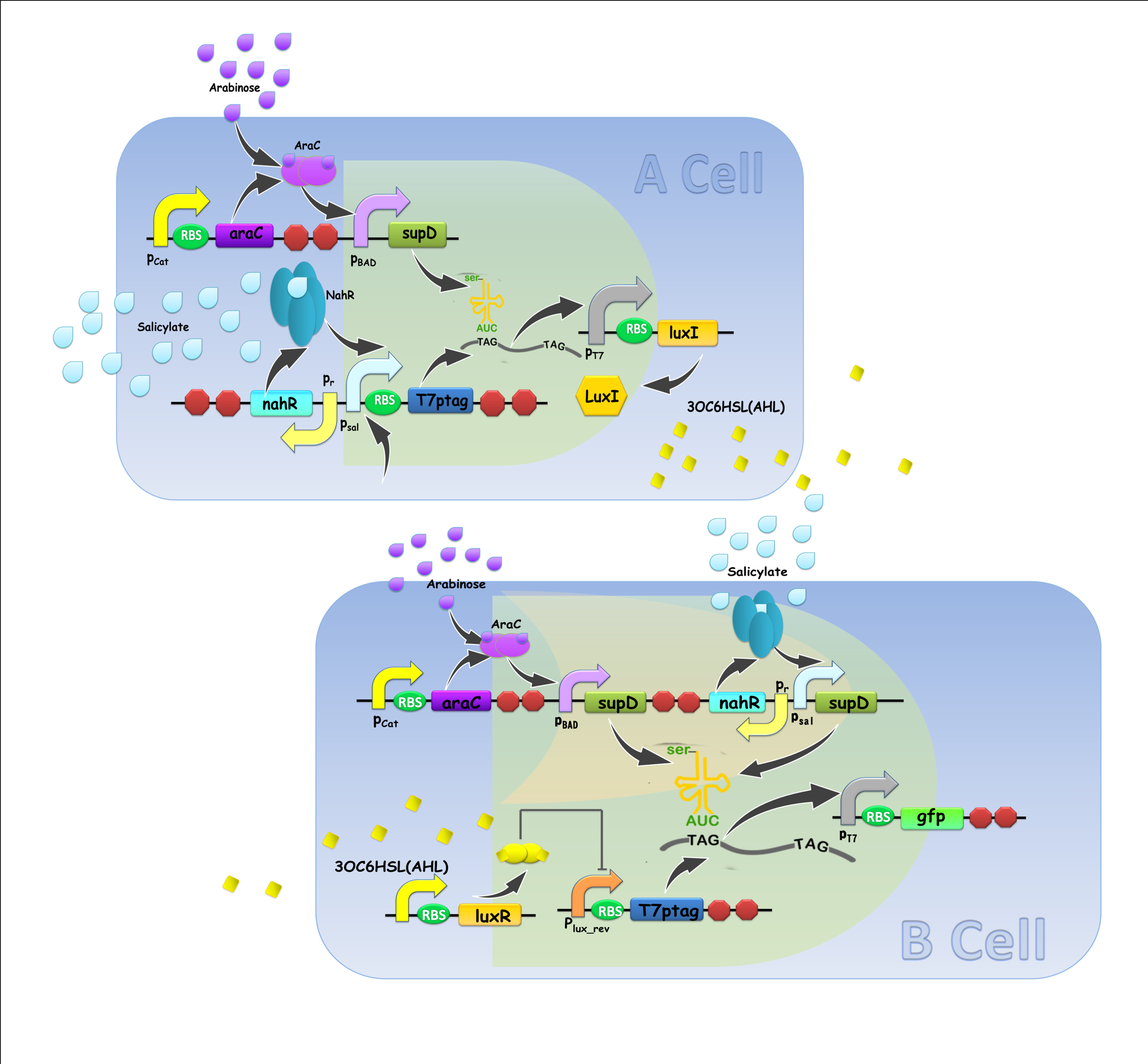
 "
"