Team:USTC-China/Project/results
From 2011.igem.org
(→The Experimente Results of the Original Design) |
|||
| (126 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{Team:USTC-China/temp}} | {{Team:USTC-China/temp}} | ||
{{Team:USTC-China/temp2}} | {{Team:USTC-China/temp2}} | ||
| + | <h1>Experiment results of the Basic Design <br/><br/>and Discussion</h1> | ||
| + | <html ><a href= "#Verification of ΔcheZ strain"><u>1.Verification of ΔcheZ strain<html></u></a> <br/> | ||
| + | <html><a href= "#Verification of the function of the constructed Aptamer-cheZ part"> <u>2.Verification of the function of the constructed Aptamer-cheZ part</u></a></html><br/> | ||
| + | <html><a href= "#Verification of the original Toggle-switch from PKU"> <u>3.Verification of the original Toggle-switch from PKU<html></u> </a> <br/> | ||
| + | <html><a href= "#Verification of the modified Toggle-switch"> <u>4.Verification of the modified Toggle-switch</u> </a> </html><br/> | ||
| + | <html><a href= "#Test of Incorporated Aptamer-cheZ part into one side of the original Toggle-switch"> <u>5.Test of Incorporated Aptamer-cheZ part into one side of the original Toggle-switch<html></u> </a><br/> | ||
| + | <html><a href= "#Test of Incorporated Aptamer-cheZ part into one side of the modified Toggle-switch"> <u>6.Test of Incorporated Aptamer-cheZ part into one side of the modified Toggle-switch</u> </a> </html><br/> | ||
| - | |||
| - | + | <html><a name="Verification of ΔcheZ strain" id=""></a></html> | |
| + | |||
| + | ==Verification of ΔcheZ strain== | ||
<p> The size of colonies of ''E.coli'' strain RP1616 was much smaller than that ''E.coli'' steain RP437 under the same circumstance and after same period of incubation time(about 10h), and the result is shown in Figure1. In colony PCR using the primers of cheZ gene following , RP437 absolutely has much more outcomes than RP1616 and we conclude that RP1616 is actually a ΔcheZ strain.</p> | <p> The size of colonies of ''E.coli'' strain RP1616 was much smaller than that ''E.coli'' steain RP437 under the same circumstance and after same period of incubation time(about 10h), and the result is shown in Figure1. In colony PCR using the primers of cheZ gene following , RP437 absolutely has much more outcomes than RP1616 and we conclude that RP1616 is actually a ΔcheZ strain.</p> | ||
| - | + | <html> | |
| - | <p align=center>Figure1.The result of colony PCR(From left to right, the first lane is the marker, the third and the forth lane is the PCR outcome of strain RP437 and strain RP1616, the sixth and the seventh lane is the PCR outcome of strain RP437 and strain RP1616.)</p> | + | <div class="center"><div class="thumb tnone" style="width:352px;"><div class="thumbinner" style="width:352px;"><a href="/File:Sst().jpg" class="image"><img alt="" src="/wiki/images/thumb/4/4e/Sst%28%29.jpg/350px-Sst%28%29.jpg" width="350" height="232" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Sst().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div><p align=center class="ppp">Figure1.The result of colony PCR(From left to right, the first lane is the marker, the third and the forth lane is the PCR outcome of strain RP437 and strain RP1616, the sixth and the seventh lane is the PCR outcome of strain RP437 and strain RP1616.)</p></div></div></div></div> |
| + | </html> | ||
| + | |||
| + | <html><a name="Verification of the function of the constructed Aptamer-cheZ part" id=""></a></html> | ||
| + | |||
| + | ==Verification of the function of the constructed Aptamer-cheZ part<html><a href= "https://2011.igem.org/Team:USTC-China/Wet_Lab/protocol#Aptamer-cheZ"><font size="4"><u>(protocol1)</u></font></a><a href= "https://2011.igem.org/Team:USTC-China/Wet_Lab/protocol#Dose-Dependent"><font size="4"><u>(protocol2)</u></font></a> </html>== | ||
| + | <p> From the results shown in Figure2. We can be sure that the Aptamer-cheZ part actually works effectively, especially on 0.3% Semi-solid medium.</p> | ||
| + | <html> | ||
| + | <div class="center"><div class="thumb tnone" style="width:652px;"><div class="thumbinner" style="width:652px;"><a href="/File:X().jpg" class="image"><img alt="X().jpg" src="/wiki/images/thumb/1/15/X%28%29.jpg/650px-X%28%29.jpg" width="650" height="182" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:X().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure2. The growing state of the reprogrammed bacteria with Aptamer-cheZ part(Left:0.3%agar with 0mM Theophylline, Right:0.3%agar with 0.25mM Theophylline)</div></div></div></div> | ||
| + | </html> | ||
| + | <p align=center class="ppp"></p> | ||
| + | <html><a name="Verification of the original Toggle-switch from PKU" id=""></a></html> | ||
| + | |||
| + | ==Verification of the original Toggle-switch from PKU<html><a href= "https://2011.igem.org/Team:USTC-China/Wet_Lab/protocol#Toggle Switch"><font size="4"><u>(protocol)</u></font></a> </html>== | ||
| + | <html> | ||
| + | <div class="center"><div class="thumb tnone" style="width:302px;"><div class="thumbinner" style="width:302px;"><a href="/File:Res14.jpg" class="image"><img alt="Res14.jpg" src="/wiki/images/thumb/f/fc/Res14.jpg/300px-Res14.jpg" width="300" height="202" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Res14.jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure3.A conlony of the bacteria with the original Toggle Switch exhibit two different states(4X Objective) </div></div></div></div> | ||
| + | </html> | ||
| + | <p align=center class="ppp"></p> | ||
| + | <p> By calculating with the help of the fluorescence microscope, the average ratio between the numbers of colonies with RFP and colonies with GFP ≈ 8:25. It means the original Toggle Switch is not fit for our purpose, so we modify the original by luxPR-cI device.</p> | ||
| + | <html><a name="Verification of the modified Toggle-switch" id=""></a></html> | ||
| + | |||
| + | ==Verification of the modified Toggle-switch <html><a href= "https://2011.igem.org/Team:USTC-China/Wet_Lab/protocol#Toggle Switch"><font size="4"><u>(protocol)</u></font></a> </html>== | ||
| + | <html> | ||
| + | <div class="center"><div class="thumb tnone" style="width:552px;"><div class="thumbinner" style="width:552px;"><a href="/File:Rs14.jpg" class="image"><img alt="Rs14.jpg" src="/wiki/images/thumb/9/9a/Rs14.jpg/550px-Rs14.jpg" width="550" height="202" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Rs14.jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure4. The conlonies of the bacteria with the modified Toggle Switch exhibit two different states or just one state(4X Objective)</div></div></div></div> | ||
| + | </html> | ||
| + | <p align=center class="ppp"></p> | ||
| + | <p> By calculating with the help of the fluorescence microscope, the ratio between the numbers of colonies with RFP and colonies with GFP ≈ 6:1. It is useful to our project design.</p> | ||
| + | <html><a name="Test of Incorporated Aptamer-cheZ part into one side of the original Toggle-switch" id=""></a></html> | ||
| + | |||
| + | ==Test of Incorporated Aptamer-cheZ part into one side of the original Toggle-switch <html><a href= "https://2011.igem.org/Team:USTC-China/Wet_Lab/protocol#Semi-Solid"><font size="4"><u>(protocol1)</u></font></a> <a href= "https://2011.igem.org/Team:USTC-China/Wet_Lab/protocol#Toggle"><font size="4"><u>(protocol2)</u></font></a></html>== | ||
| + | <html> | ||
| + | <div class="center"><div class="thumb tnone" style="width:652px;"><div class="thumbinner" style="width:652px;"><a href="/File:Resul.jpg" class="image"><img alt="Resul.jpg" src="/wiki/images/thumb/f/fc/Resul.jpg/650px-Resul.jpg" width="650" height="236" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Resul.jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure5.The growing state of the reprogrammed bacteria with original Toggle-switch-Aptamer-cheZ Device(Red circle:the range of motion, Blue Triangle:the gradient of thepphylline)</div></div></div></div> | ||
| + | </html> | ||
| + | <p align=center class="ppp"></p> | ||
| + | <html> | ||
| + | <div class="center"><div class="thumb tnone" style="width:302px;"><div class="thumbinner" style="width:302px;"><a href="/File:Resddul.jpg" class="image"><img alt="Resddul.jpg" src="/wiki/images/thumb/d/de/Resddul.jpg/300px-Resddul.jpg" width="300" height="224" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Resddul.jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure6.The fluorescent Photo(4X Objective) of the colony shown in Figure5 Left.</div></div></div></div> | ||
| + | </html> | ||
| + | <p align=center class="ppp"></p> | ||
| + | <p> According to the results above, by using this device we can not make the reprogrammed bacteria be divided into two different states effectively, and because of the bias to the green fluorescence and moving towards the high concentration of the theophylline, the bacteria trending to stay are very rare.</p> | ||
| + | <html><a name="Test of Incorporated Aptamer-cheZ part into one side of the modified Toggle-switch" id=""></a></html> | ||
| + | |||
| + | ==Test of Incorporated Aptamer-cheZ part into one side of the modified Toggle-switch <html><a href= "https://2011.igem.org/Team:USTC-China/Wet_Lab/protocol#Toggle Switch-Aptamer-cheZ"><font size="4"><u>(protocol)</u></font></a> </html>== | ||
| + | <html> | ||
| + | <div class="center"><div class="thumb tnone" style="width:482px;"><div class="thumbinner" style="width:482px;"><a href="/File:Re.jpg" class="image"><img alt="Re.jpg" src="/wiki/images/thumb/c/c8/Re.jpg/480px-Re.jpg" width="480" height="137" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Re.jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure7. The fluctuation of expression of the modified Toggle Switch device, the number of bacteria with red fluorescent decreases from right to left.</div></div></div></div> | ||
| + | </html> | ||
| - | == | + | <p> </p> |
| - | + | <html> | |
| - | + | <div class="center"><div class="thumb tnone" style="width:902px;"><div class="thumbinner" style="width:902px;"><a href="/File:Re3.jpg" class="image"><img alt="Re3.jpg" src="/wiki/images/thumb/a/a4/Re3.jpg/900px-Re3.jpg" width="900" height="162" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Re3.jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure8. Four different growing states of the reprogrammed bacteria, in which the yellow circle represent the range of motion of reprogrammed bacteria(from the second tube(from left to right) shown in Figure7), and from left to right the concentration of the theophylline increases.</div></div></div></div> | |
| - | + | </html> | |
| - | < | + | |
| - | + | ||
| - | + | ||
| - | == | + | <p> </p> |
| - | | + | <html> |
| - | < | + | <div class="center"><div class="thumb tnone" style="width:452px;"><div class="thumbinner" style="width:452px;"><a href="/File:Results12.jpg" class="image"><img alt="Results12.jpg" src="/wiki/images/thumb/c/cb/Results12.jpg/450px-Results12.jpg" width="450" height="337" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Results12.jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure9.The fluorescent Photo(4X Objective)of the colony1 in the fourth image of Figure8, and from left to right the concentration of the theophylline increases.</div></div></div></div> |
| + | </html> | ||
| + | <p align=center class="ppp"></p> | ||
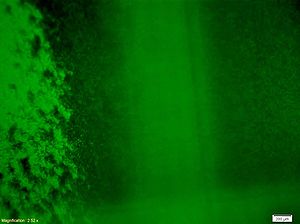
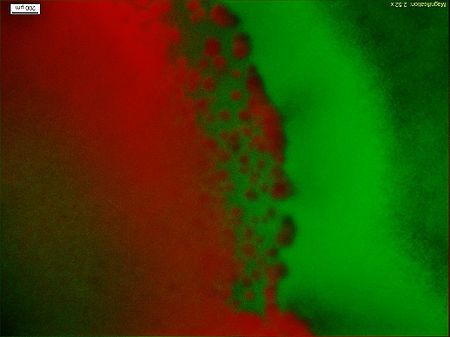
| + | <p> Figure9 clearly display the dividing line between the reprogrammed bacteria at two different states, and the bacteria mainly express GFP indeed moving towards the high concentration of theophylline. This means the modified Toggle-switch-Aptamer-cheZ actually work as our design.</p> | ||
| + | <html> | ||
| + | <div class="center"><div class="thumb tnone" style="width:502px;"><div class="thumbinner" style="width:502px;"><a href="/File:Dddw().jpg" class="image"><img alt="Dddw().jpg" src="/wiki/images/thumb/9/9c/Dddw%28%29.jpg/500px-Dddw%28%29.jpg" width="500" height="297" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Dddw().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure10.The SDS-PAGE result of cheZ proteins in the bacteria from B1 to B5 (Blue Triangle:the gradient of thepphylline)</div></div></div></div> | ||
| + | </html> | ||
| - | == | + | <p align=center class="ppp"> </p> |
| - | + | <html> | |
| - | < | + | <div class="center"><div class="thumb tnone" style="width:502px;"><div class="thumbinner" style="width:502px;"><a href="/File:Dddk().jpg" class="image"><img alt="Dddk().jpg" src="/wiki/images/thumb/4/40/Dddk%28%29.jpg/500px-Dddk%28%29.jpg" width="500" height="117" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Dddk().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure11.The basic parameters of the SDS-PAGE</div></div></div></div> |
| + | </html> | ||
| - | + | <p align=center class="ppp"></p> | |
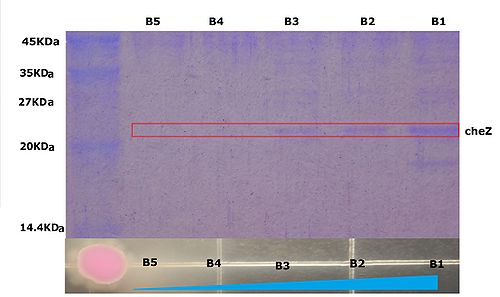
| - | | + | <p> Figure10 shows that the expression of cheZ of the bacteria increases along the theophylline gradient, which further prove the results shown before. </p> |
| - | == | + | ==Discussion== |
| - | + | <p> The experiment result mainly depend on the the random fluctuation of expression of the modified Toggle Switch device, so this device has a certain probability to switch from one state to the other, then it may lead to stopping of the moving bacteria like colony2(Figure12.).</P> | |
| - | + | <html> | |
| - | + | <div class="center"><div class="thumb tnone" style="width:352px;"><div class="thumbinner" style="width:352px;"><a href="/File:R2.jpg" class="image"><img alt="R2.jpg" src="/wiki/images/thumb/f/fb/R2.jpg/350px-R2.jpg" width="350" height="262" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:R2.jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure12.The fluorescent Photo(4X Objective)of the colony2 in the lower right of Figure8, and from left to right the concentration of the theophylline increases.</div></div></div></div> | |
| - | + | </html> | |
| - | | + | <p align=center class="ppp"></p> |
| + | <p> To weaken this randomness, we may use the quorum sensing to control the moving of the reprogrammed bacteria, and we construct a model to simulate this proposal by computer ([https://2011.igem.org/Team:USTC-China/Drylab/modeling '''Modeling''']). | ||
Latest revision as of 18:35, 28 October 2011

Experiment results of the Basic Design
and Discussion
1.Verification of ΔcheZ strain
2.Verification of the function of the constructed Aptamer-cheZ part
3.Verification of the original Toggle-switch from PKU
4.Verification of the modified Toggle-switch
5.Test of Incorporated Aptamer-cheZ part into one side of the original Toggle-switch
6.Test of Incorporated Aptamer-cheZ part into one side of the modified Toggle-switch
Verification of ΔcheZ strain
The size of colonies of E.coli strain RP1616 was much smaller than that E.coli steain RP437 under the same circumstance and after same period of incubation time(about 10h), and the result is shown in Figure1. In colony PCR using the primers of cheZ gene following , RP437 absolutely has much more outcomes than RP1616 and we conclude that RP1616 is actually a ΔcheZ strain.
Verification of the function of the constructed Aptamer-cheZ part(protocol1)(protocol2)
From the results shown in Figure2. We can be sure that the Aptamer-cheZ part actually works effectively, especially on 0.3% Semi-solid medium.
Verification of the original Toggle-switch from PKU(protocol)
By calculating with the help of the fluorescence microscope, the average ratio between the numbers of colonies with RFP and colonies with GFP ≈ 8:25. It means the original Toggle Switch is not fit for our purpose, so we modify the original by luxPR-cI device.
Verification of the modified Toggle-switch (protocol)
By calculating with the help of the fluorescence microscope, the ratio between the numbers of colonies with RFP and colonies with GFP ≈ 6:1. It is useful to our project design.
Test of Incorporated Aptamer-cheZ part into one side of the original Toggle-switch (protocol1) (protocol2)
According to the results above, by using this device we can not make the reprogrammed bacteria be divided into two different states effectively, and because of the bias to the green fluorescence and moving towards the high concentration of the theophylline, the bacteria trending to stay are very rare.
Test of Incorporated Aptamer-cheZ part into one side of the modified Toggle-switch (protocol)
Figure9 clearly display the dividing line between the reprogrammed bacteria at two different states, and the bacteria mainly express GFP indeed moving towards the high concentration of theophylline. This means the modified Toggle-switch-Aptamer-cheZ actually work as our design.
Figure10 shows that the expression of cheZ of the bacteria increases along the theophylline gradient, which further prove the results shown before.
Discussion
The experiment result mainly depend on the the random fluctuation of expression of the modified Toggle Switch device, so this device has a certain probability to switch from one state to the other, then it may lead to stopping of the moving bacteria like colony2(Figure12.).
To weaken this randomness, we may use the quorum sensing to control the moving of the reprogrammed bacteria, and we construct a model to simulate this proposal by computer (Modeling).
 "
"