Team:HKU-Hong Kong/Lab Diaries
From 2011.igem.org
(Difference between revisions)
| (3 intermediate revisions not shown) | |||
| Line 5: | Line 5: | ||
|style="width:900px;"|'''Week 1''' | |style="width:900px;"|'''Week 1''' | ||
Transformation of reporter DNA (pEGFP-loxp-km-loxp) into DH10B (non-virulent strain E. coli) with antibiotic resistance (Chloramphenicol – Cm) | Transformation of reporter DNA (pEGFP-loxp-km-loxp) into DH10B (non-virulent strain E. coli) with antibiotic resistance (Chloramphenicol – Cm) | ||
| - | |||
|- | |- | ||
|style="width:900px;"|'''Week 2''' | |style="width:900px;"|'''Week 2''' | ||
| Line 24: | Line 23: | ||
<LI>2uL of 10-fold diluted pEGFP-loxp-km-loxp- tetO2 – 3 was transformed into DH10B to greatly amplify the product by using bacterial cells. | <LI>2uL of 10-fold diluted pEGFP-loxp-km-loxp- tetO2 – 3 was transformed into DH10B to greatly amplify the product by using bacterial cells. | ||
</OL> | </OL> | ||
| - | |||
|- | |- | ||
|style="width:900px;"|'''Week 3''' | |style="width:900px;"|'''Week 3''' | ||
| Line 58: | Line 56: | ||
</OL> | </OL> | ||
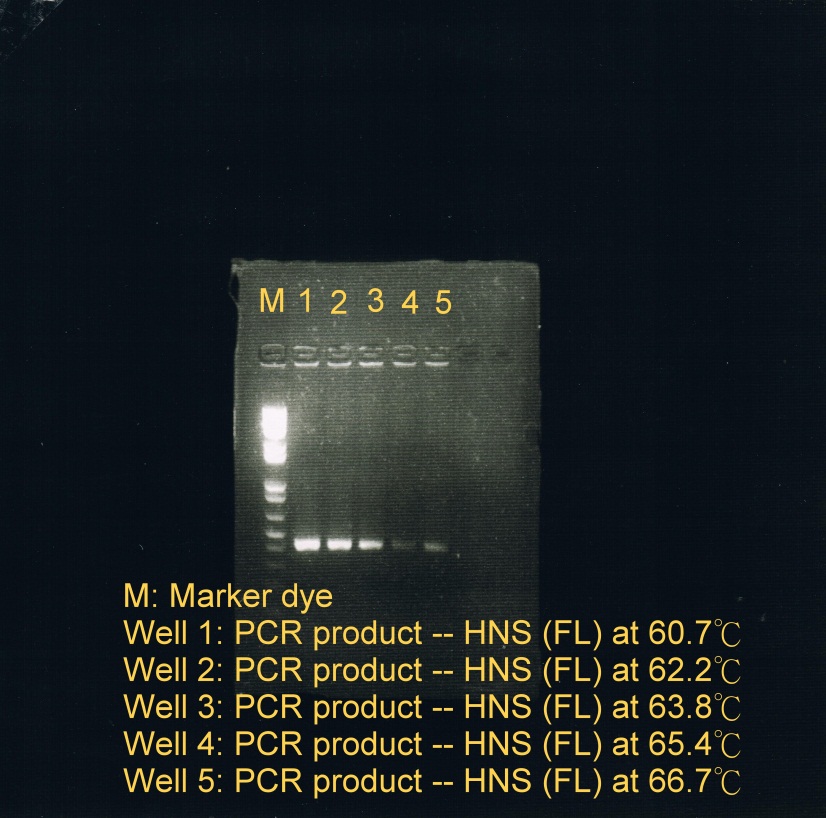
<LI>Electrophoresis was carried to determine which temperature best suit the annealing phase. From the gel image, it was observed that a high concentration of products was resulted at annealing temperature between 60.7C and 62.2C. | <LI>Electrophoresis was carried to determine which temperature best suit the annealing phase. From the gel image, it was observed that a high concentration of products was resulted at annealing temperature between 60.7C and 62.2C. | ||
| + | |||
<div ALIGN=CENTER> | <div ALIGN=CENTER> | ||
{| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | {| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| Line 66: | Line 65: | ||
|} | |} | ||
</div> | </div> | ||
| - | |||
|- | |- | ||
|style="width:900px;"|'''Week 4-5''' | |style="width:900px;"|'''Week 4-5''' | ||
| Line 73: | Line 71: | ||
<LI>Reverse PCR was used to insert tetO2 -0 and tetO2 – 4 into pEGFP-loxp-km-loxp. | <LI>Reverse PCR was used to insert tetO2 -0 and tetO2 – 4 into pEGFP-loxp-km-loxp. | ||
<LI>2uL of pEGFP-loxp-km-loxp- tetO2 – 0 and pEGFP-loxp-km-loxp- tetO2 – 4 were transformed into DH10B separately to greatly amplify the product by using bacterial cells. | <LI>2uL of pEGFP-loxp-km-loxp- tetO2 – 0 and pEGFP-loxp-km-loxp- tetO2 – 4 were transformed into DH10B separately to greatly amplify the product by using bacterial cells. | ||
| - | |||
|- | |- | ||
|style="width:900px;"|'''Week 6''' | |style="width:900px;"|'''Week 6''' | ||
| Line 92: | Line 89: | ||
</OL> | </OL> | ||
</OL> | </OL> | ||
| - | |||
|- | |- | ||
|style="width:900px;"|'''Week 7''' | |style="width:900px;"|'''Week 7''' | ||
| Line 98: | Line 94: | ||
<OL> | <OL> | ||
<LI>The sticky ends of the enzyme products was transformed into blunt ends using PCR. | <LI>The sticky ends of the enzyme products was transformed into blunt ends using PCR. | ||
| - | |||
|- | |- | ||
|style="width:900px;"|'''Week 8''' | |style="width:900px;"|'''Week 8''' | ||
| Line 115: | Line 110: | ||
<OL> | <OL> | ||
<LI>0.5uL of CIP alkaline phosphatase was added to prevent self-ligation to each enzyme digest reaction mixture. | <LI>0.5uL of CIP alkaline phosphatase was added to prevent self-ligation to each enzyme digest reaction mixture. | ||
| - | |||
|- | |- | ||
|style="width:900px;"|'''Week 9''' | |style="width:900px;"|'''Week 9''' | ||
| Line 124: | Line 118: | ||
</OL> | </OL> | ||
<LI>Production of tetO2 – 0 –sfGFP | <LI>Production of tetO2 – 0 –sfGFP | ||
| + | |||
<div ALIGN=CENTER> | <div ALIGN=CENTER> | ||
{| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | {| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| Line 132: | Line 127: | ||
|} | |} | ||
</div> | </div> | ||
| - | |||
|- | |- | ||
|style="width:900px;"|'''Week 10''' | |style="width:900px;"|'''Week 10''' | ||
| Line 151: | Line 145: | ||
<LI>Fluorescence intensity were checked at OD600 using spectrophotometer | <LI>Fluorescence intensity were checked at OD600 using spectrophotometer | ||
</OL> | </OL> | ||
| - | |||
|- | |- | ||
|style="width:900px;"|'''Week 11''' | |style="width:900px;"|'''Week 11''' | ||
| Line 173: | Line 166: | ||
<LI>pSB1C3-HNS | <LI>pSB1C3-HNS | ||
<LI>Electrophoresis was carried out to confirm the success of extraction) (except J23116RH) | <LI>Electrophoresis was carried out to confirm the success of extraction) (except J23116RH) | ||
| + | |||
| + | <div ALIGN=CENTER> | ||
| + | {| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:Week_11.jpg|250px]] | ||
| + | |- | ||
| + | |Confirmation of successful extraction | ||
| + | |} | ||
| + | </div> | ||
| + | |||
| + | <div ALIGN=CENTER> | ||
| + | {| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:Week_11_(2).jpg|250px]] | ||
| + | |- | ||
| + | |Confirmation of successful extraction | ||
| + | |} | ||
| + | </div> | ||
</OL> | </OL> | ||
<LI>Purification of NdeI digested pET28a | <LI>Purification of NdeI digested pET28a | ||
| - | |||
|- | |- | ||
|style="width:900px;"|'''Week 12''' | |style="width:900px;"|'''Week 12''' | ||
| Line 191: | Line 201: | ||
<LI>tetO2 – 1 | <LI>tetO2 – 1 | ||
<LI>sfGFP | <LI>sfGFP | ||
| - | |||
|- | |- | ||
|style="width:900px;"|'''Week 13''' | |style="width:900px;"|'''Week 13''' | ||
| Line 212: | Line 221: | ||
<LI>J23103RH | <LI>J23103RH | ||
<LI>J23116R | <LI>J23116R | ||
| + | |- | ||
| + | |style="width:900px;"|'''Week 14''' | ||
| + | <OL> | ||
| + | <LI>Production of J23116-tetR::HNS (2-90) | ||
| + | <OL> | ||
| + | <LI>Colony PCR was used to confirm the success of production | ||
| + | </OL> | ||
| + | <LI>Fluorescence of our samples were tested [in M1655 (with tetO-0, tetO-1 or tetO-0-sfGFP)] | ||
| + | <OL> | ||
| + | <LI>J23109R | ||
| + | <LI>J23109RH | ||
| + | <LI>J23109RHH | ||
| + | <LI>J23116R | ||
| + | <LI>J23116RHH | ||
| + | </OL> | ||
| + | <LI>Check for the OD488 to OD510 (green fluorescence); OD600 (cell concentration in the solution) | ||
| + | <OL> | ||
| + | <LI>With 1 set for CTC | ||
| + | <OL> | ||
| + | <LI>Procedures | ||
| + | <OL> | ||
| + | <LI>A colony of the above samples was incubated in 3mL LB broth with antibiotics overnight. | ||
| + | <LI>1:50 diluted in M9 solution (10mL) (for CTC set: add CTC 20ug/mL). | ||
| + | <LI>Samples were then incubated at 37C for 6 hours in dark. | ||
| + | <LI>Each sample was then concentrated to 200uL. | ||
| + | <LI>Samples were loaded into a 96-well plate to check for OD using spectrophotometer. | ||
Latest revision as of 06:59, 5 October 2011
| Lab Diaries |
| Week 1
Transformation of reporter DNA (pEGFP-loxp-km-loxp) into DH10B (non-virulent strain E. coli) with antibiotic resistance (Chloramphenicol – Cm) |
Week 2
|
Week 3
|
| Week 4-5
tetO2 – 0 and tetO2 – 4
|
| Week 6
Overlap PCR was carried out to produce fusion protein gene
|
| Week 7
Overlap PCR was carried out to produce fusion protein gene tetR::HNS (2-90)
|
Week 8
|
Week 9
|
Week 10
|
Week 11
|
Week 12
|
Week 13
|
Week 14
|
 "
"