Team:HKU-Hong Kong/Lab Protocol
From 2011.igem.org
(Difference between revisions)
| (20 intermediate revisions not shown) | |||
| Line 74: | Line 74: | ||
<div ALIGN=CENTER> | <div ALIGN=CENTER> | ||
{| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | {| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
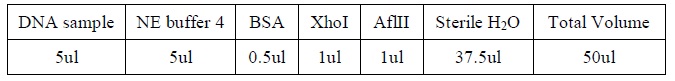
| - | |[[Image:Table_1.jpg| | + | |- |
| - | + | |[[Image:Table_1.jpg|500px]] | |
| + | |- | ||
| + | |Reagents for DNA digestion. | ||
| + | |} | ||
</div> | </div> | ||
| - | + | <LI>All steps should be carried out on ice. | |
| - | + | <LI>Mix well after addition of all the reagent. | |
| - | + | <LI>Incubate the mixture at 37oC for several hours. | |
| - | + | ||
| - | |||
| - | |||
| - | |||
|- | |- | ||
|style="width:900px;font-size: 1.3em;"|Miniprep(Adopt from Qiagen) | |style="width:900px;font-size: 1.3em;"|Miniprep(Adopt from Qiagen) | ||
| Line 111: | Line 110: | ||
|- | |- | ||
|style="width:900px;| | |style="width:900px;| | ||
| - | + | <OL> | |
| - | Colony PCR | + | <LI>Colony PCR |
| - | + | <OL> | |
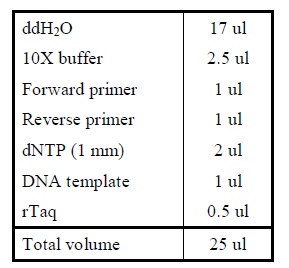
| - | + | <LI>Add the following reagents into a PCR tube (in order) and mix well. | |
| - | + | ||
<div ALIGN=CENTER> | <div ALIGN=CENTER> | ||
{| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | {| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| - | |[[Image:Colony_PCR.jpg| | + | |- |
| - | + | |[[Image:Colony_PCR.jpg|225px]] | |
| + | |- | ||
| + | |Reagents for colony PCR. | ||
| + | |} | ||
</div> | </div> | ||
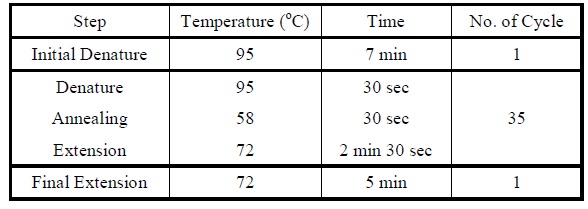
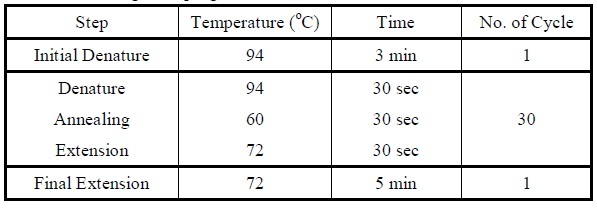
| - | + | <LI>Set the following PCR program. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<div ALIGN=CENTER> | <div ALIGN=CENTER> | ||
{| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | {| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| - | |[[Image:Colony_pcr_program.jpg| | + | |- |
| - | + | |[[Image:Colony_pcr_program.jpg|475px]] | |
| + | |- | ||
| + | |Reaction program for colony PCR. | ||
| + | |} | ||
</div> | </div> | ||
| - | + | </OL> | |
| - | + | ||
| - | + | ||
| - | + | <LI>Reverse PCR | |
| - | Reverse PCR | + | <OL> |
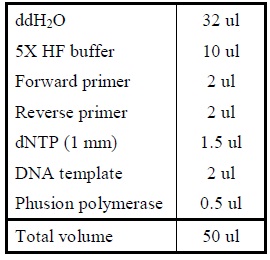
| - | + | <LI>Add the following reagents into a PCR tube (in order) and mix well. | |
<div ALIGN=CENTER> | <div ALIGN=CENTER> | ||
{| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | {| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| - | |[[Image: | + | |- |
| - | + | |[[Image:Reverse_pcr.jpg|225px]] | |
| + | |- | ||
| + | |Reagents for reverse PCR. | ||
| + | |} | ||
</div> | </div> | ||
| - | + | <LI>Set the following PCR program. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<div ALIGN=CENTER> | <div ALIGN=CENTER> | ||
{| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | {| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| - | |[[Image:Reverse_pcr_program.jpg| | + | |- |
| - | + | |[[Image:Reverse_pcr_program.jpg|475px]] | |
| + | |- | ||
| + | |Reaction program for reverse PCR. | ||
| + | |} | ||
</div> | </div> | ||
| - | + | </OL> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | Overlap PCR | + | <LI>Overlap PCR |
<OL> | <OL> | ||
| - | <LI>First, two PCR reactions are set for amplifying the two genes, tetR and HNS, separately. | + | <LI>First, two PCR reactions are set for amplifying the two genes, tetR and HNS, separately. |
| - | <LI>Add the following reagents into a PCR tube (in order) and mix well.</ | + | <LI>Add the following reagents into a PCR tube (in order) and mix well. |
| - | <LI>Set the following PCR program.</ | + | <div ALIGN=CENTER> |
| - | <LI>Set up another PCR reaction using a primer with a linker to link the two genes. | + | {| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; |
| - | <LI>Add the following reagents into a PCR tube (in order) and mix well.</ | + | |- |
| - | <LI>Set the following PCR program.</ | + | |[[Image:Overlap_pcr.jpg|500px]] |
| - | <LI>Set up another PCR reaction to further amplify the fused product. | + | |- |
| - | <LI>Add the following reagents into a PCR tube (in order) and mix well.</ | + | |Reagents for amplifying genes. |
| - | <LI>Set the following PCR program. | + | |} |
| - | < | + | </div> |
| - | </ | + | <LI>Set the following PCR program. |
| + | <div ALIGN=CENTER> | ||
| + | {| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:Overlap_pcr_program.jpg|500px]] | ||
| + | |- | ||
| + | |Reaction program for amplifying genes. | ||
| + | |} | ||
| + | </div> | ||
| + | <LI>Set up another PCR reaction using a primer with a linker to link the two genes. | ||
| + | <LI>Add the following reagents into a PCR tube (in order) and mix well. | ||
| + | <div ALIGN=CENTER> | ||
| + | {| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:Overlap_pcr_program.jpg|500px]] | ||
| + | |- | ||
| + | |Reagents for linking two genes. | ||
| + | |} | ||
| + | </div> | ||
| + | <LI>Set the following PCR program. | ||
| + | <div ALIGN=CENTER> | ||
| + | {| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:Overlap_pcr_program_2.jpg|500px]] | ||
| + | |- | ||
| + | |Reaction program for linking two genes. | ||
| + | |} | ||
| + | </div> | ||
| + | <LI>Set up another PCR reaction to further amplify the fused product. | ||
| + | <LI>Add the following reagents into a PCR tube (in order) and mix well. | ||
| + | <div ALIGN=CENTER> | ||
| + | {| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:Overlap_pcr_3.jpg|300px]] | ||
| + | |- | ||
| + | |Reagents for amplifying the fused product. | ||
| + | |} | ||
| + | </div> | ||
| + | <LI>Set the following PCR program. | ||
| + | <div ALIGN=CENTER> | ||
| + | {| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:Overlap_pcr_program_3.jpg|500px]] | ||
| + | |- | ||
| + | |Reaction program for amplifying the fused product. | ||
| + | |} | ||
| + | </div> | ||
| + | |||
|- | |- | ||
| Line 178: | Line 221: | ||
|style="width:900px;| | |style="width:900px;| | ||
<OL> | <OL> | ||
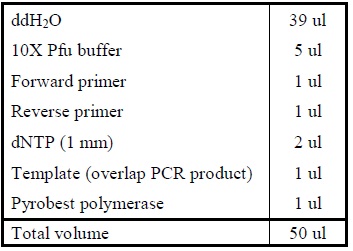
| - | <LI>Add the following reagents, with the enzymes added at the last, into a tube.</ | + | <LI>Add the following reagents, with the enzymes added at the last, into a tube. |
| - | <LI>Incubate at 16oC overnight. | + | <div ALIGN=CENTER> |
| + | {| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:DNA_ligation.jpg|550px]] | ||
| + | |- | ||
| + | |Composition for DNA ligation. | ||
| + | |} | ||
| + | </div> | ||
| + | <LI>Incubate at 16oC overnight. | ||
</OL> | </OL> | ||
| Line 325: | Line 376: | ||
|style="width:900px;| | |style="width:900px;| | ||
<OL> | <OL> | ||
| - | <LI>Gel Preparation | + | <LI>Gel Preparation |
<OL> | <OL> | ||
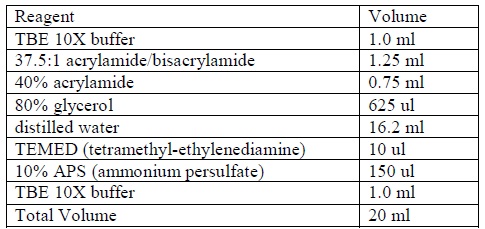
| - | <LI>Clean all the glassware by distilled water (ions-free). | + | <LI>Clean all the glassware by distilled water (ions-free). |
| - | <LI>Prepare a non-denaturing 4% acrylamide gel according to the following formula:</ | + | <LI>Prepare a non-denaturing 4% acrylamide gel according to the following formula: |
| - | <LI>Allow the gel to stand until the gel is completely polymerized. | + | <div ALIGN=CENTER> |
| + | {| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:Gel_shift_assay_gel_preparation.jpg|400px]] | ||
| + | |- | ||
| + | |Formula for preparing acrylamide gel. | ||
| + | |} | ||
| + | </div> | ||
| + | <LI>Allow the gel to stand until the gel is completely polymerized. | ||
</OL> | </OL> | ||
| - | <LI>DNA Binding Reactions | + | |
| + | <LI>DNA Binding Reactions | ||
<OL> | <OL> | ||
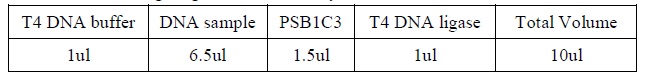
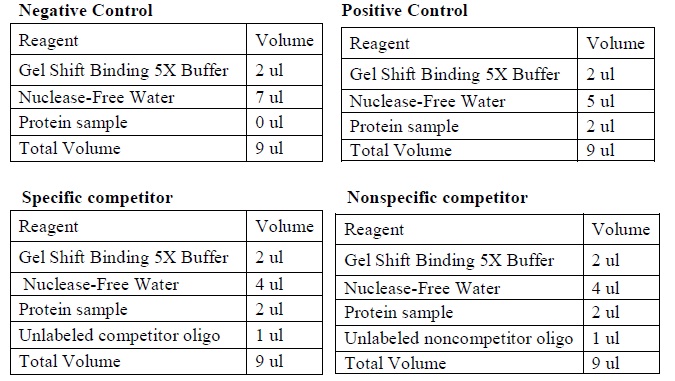
| - | <LI>Set up four binding reactions (if necessary) with the following composition.</ | + | <LI>Set up four binding reactions (if necessary) with the following composition. |
| - | <LI>Incubate the reactions at room temperature for 10 minutes. | + | <div ALIGN=CENTER> |
| - | <LI>Add 1 ul of labeled DNA sample to each reaction. | + | {| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; |
| - | <LI>Incubate the reactions at room temperature for 20 minutes. | + | |- |
| - | <LI>Add 1 ul of 10X loading buffer for each reaction. | + | |[[Image:Gel_shoft_assay_DNA_binding_reactions.jpg|500px]] |
| + | |- | ||
| + | |The four binding reactions: Negative control, Positive control, Specific competitor and Non-specific competitor | ||
| + | |} | ||
| + | </div> | ||
| + | <LI>Incubate the reactions at room temperature for 10 minutes. | ||
| + | <LI>Add 1 ul of labeled DNA sample to each reaction. | ||
| + | <LI>Incubate the reactions at room temperature for 20 minutes. | ||
| + | <LI>Add 1 ul of 10X loading buffer for each reaction. | ||
</OL> | </OL> | ||
| - | <LI>Electrophoresis | + | |
| + | <LI>Electrophoresis | ||
<OL> | <OL> | ||
| - | <LI>Pre-run the gel in 0.5X TBE buffer for 10 minutes at 350V. | + | <LI>Pre-run the gel in 0.5X TBE buffer for 10 minutes at 350V. |
| - | <LI>Load the sample. | + | <LI>Load the sample. |
| - | <LI>Run the gel at 350V until the loading dye reached three fourth of the gel. | + | <LI>Run the gel at 350V until the loading dye reached three fourth of the gel. |
| - | <LI>Maintain the gel temperature under 30oC. | + | <LI>Maintain the gel temperature under 30oC. |
</OL> | </OL> | ||
</OL> | </OL> | ||
| - | <LI>Protocol adopt from: http://www.promega.com/~/media/Files/Resources/Protocols/Technical%20Bulletins/0/Gel%20Shift%20Assay%20Systems.ashx | + | |
| + | <LI>Protocol adopt from: | ||
| + | |||
| + | <LI>http://www.promega.com/~/media/Files/Resources/Protocols/Technical%20Bulletins/0/Gel%20Shift%20Assay%20Systems.ashx | ||
Latest revision as of 03:19, 5 October 2011
| Lab Protocol |
| A. DNA WORK |
| Agarose Gel Electrophoresis |
|
| DNA Extraction from Agarose Gel |
|
| DNA Digestion |
|
| Miniprep(Adopt from Qiagen) |
|
| Polymerase Chain Reaction |
|
| DNA ligation |
|
| Sequencing |
|
| B. BACTERIAL WORK |
| Overnight culture |
|
| Preparation of competent cell |
|
| Spread plate |
|
| Streak plate |
|
| Transformation |
|
| C. Preparation of materials |
| Preparation of ampicillin |
|
| Preparation of LB agar plate |
|
| Preparation of LB broth |
|
| Preparation of 1X TAE buffer |
|
| D. Protein Work |
| Gel Shift Assays (Adopt from Promega) |
|
 "
"