Team:USTC-Software/tutorial
From 2011.igem.org
| (49 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
__NOTOC__ | __NOTOC__ | ||
| - | {{:Team:USTC-Software/ | + | {{:Team:USTC-Software/header}} |
| - | + | ||
<html xmlns="http://www.w3.org/1999/xhtml"> | <html xmlns="http://www.w3.org/1999/xhtml"> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<body> | <body> | ||
| - | |||
| - | + | <div id="content_wrapper"> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <div id="intro"> | |
| - | + | <p> Contents: | |
| - | + | <ul type="square"> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | <li><a href="#MoDeL Demo">MoDeL Demo</a></li> | ||
| + | <ul type="circle"> | ||
| + | <li><a href="#toggle switch">toggle switch</a></li> | ||
| + | <li><a href="#RTC two counter (RiboRegulated transcription cascade)">RTC two counter (RiboRegulated transcription cascade)</a></li> | ||
| + | </ul> | ||
| + | |||
| + | </ul> | ||
| + | |||
| + | </div> | ||
| - | |||
| - | + | <h2><a name="MoDeL Demo" id="MoDeL Demo"></a>MoDeL Demo</h2> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | <h3><a name="toggle switch" id="toggle switch"></a>Part1: toggle switch.</h3> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | </ | + | <img src="https://static.igem.org/mediawiki/2011/e/ed/USTC_Software_algo101.jpg" width="512"> |
| - | + | ||
| - | < | + | <p>Toggle switch is characterized by mutual inhibitory network. That is, the protein product of one gene represses the promoter of the other, and vice versa. So with proper parameters, the system will stay at one stable steady state at equilibrium. In other words, one gene's product will be domanant. But with proper inducers (usually IPTG or ATC), the system manage to flop from one state to another, still keeping steady.</p> |
| - | < | + | <p>We tried an instance of a toggle switch composed of a promoter which can be repressed by laci protein, and is denoted as pLac(r0010). And the other promoter is tet-repressible and be denoted by pTet(r0040). The plac promoter promotes the expression of TetR(c0040), And the LacR(c0012) is initiated by pTet(r0040).</p> |
| - | < | + | <!--<img src="https://static.igem.org/mediawiki/2011/0/06/USTC_Software_algo102.jpg">--> |
| + | </html> | ||
| + | [[File:toggleswitch_lachesis.jpg|500px]] | ||
| + | <html> | ||
| + | <p>Without inducers, which protein will dominate depend on the binding affinity of the repressor to the promoter. In this case, it's the LacR protein take control at null input of inducers. The only input to the algorithm is a input file like this.</p> | ||
| - | < | + | <img src="https://static.igem.org/mediawiki/2011/9/9d/USTC_Software_algo103.jpg"> |
| - | + | ||
| - | <p> | + | <p>Here are a bit explanations to the input file</p> |
| - | + | <p>As to the seedspecies section:<br/> | |
| + | # Medium iptg : IPTG is contained in the compartment named medium<br/> | ||
| + | # Ecoli dna1 d:r0040(tetr1,tetr2)-b0034(rib)-c0012(dna,iptg,dim)-b0014()<br/> | ||
| + | #dna1_init<br/> | ||
| + | # d:means it's a strand of dna composed of 4 parts.<br/> | ||
| + | # The promoters have two binding site(tetr1 and tetr2) for associative repressors. <br/> | ||
| + | # c0012(dna,iptg,dim) means the c0012 is a DNA sequence. Iptg means the product #of c0012, which is LacI protein, in terms of dimer, can be binded to iptg which #deactivate it's repression to placi promoter. <br/> | ||
| + | #b0014()is the terminator.<br/> | ||
| + | #dna1_init means the initial concentration of dna1 is dana1_init which is given in the parameter section of the input file.</p> | ||
| - | <p> | + | <p>Once the input file is provided, both the net and SBML file will be generated within a second. The species "discovered" by the algorithm is illustrated below, totally 18 substances in such a simple network.</p> |
| - | + | ||
| - | < | + | <img src="https://static.igem.org/mediawiki/2011/2/29/USTC_Software_algo104.jpg"> |
| - | + | <p>A bit explanations are given to the table above.</p> | |
| - | <p> | + | <p>1.<br/> |
| - | + | #nb: non-biobrick part, indicating a constraint of only one part constituting a sequence, making it significantly distinguished from DNA, RNA, Protein sequences.<br/> | |
| + | #d: means the following thing is a sequence of dna with structure information.<br/> | ||
| + | # b0014() is a terminator with no binding site.<br/> | ||
| + | # the hyphen sign connects the parts on a dna.<br/> | ||
| + | #C0012*(dim,dna,iptg) means the c0012 is a dna sequence, and the product of c0012 which is #laci protein, can form dimers, iptg can bind to monomers of laci .</p> | ||
| - | <p> | + | <p>2.<br/> |
| + | As you can see from species 12 to 15, the advantage of this approach is that it contains information about the structure of the species. For instance, s12 is nb:i0001(laci!1).p:c0012(dim!2,dna,iptg!1).p:c0012(dim!2,dna,iptg)</p> | ||
| - | + | <p>The ! is used to denote binding , here iptg bind to the laci denoted by !1, and the . is used to separate different molecules of one species. The molecule that comprise the s12 species are nb:i0001(laci!1),p:c0012(dim!2,dna,iptg!1),p:c0012(dim!2,dna,iptg).</p> | |
| - | <p>The | + | <p>The two proteins bind to form dimers through bingding !2.<br/> |
| + | So now you can imagine the structure of s12 species.<br/> | ||
| + | The network generated by our software is as below. User can drag the nodes to the place they want to have a better clarity of the network.</p> | ||
| - | < | + | <img src="https://static.igem.org/mediawiki/2011/e/e1/USTC_Software_algo105.jpg" width="644"> |
| - | + | <p>One thing to note is the assumptions made by Chen LIAO:<br/> | |
| + | 1.Both LacR protein and TetR protein can form dimers. But only the dimers, rather than the monomers, can bind to the promoter regions | ||
| + | to repress the expressions of the downstream genes.<br/> | ||
| + | <em>(#note that this a an advantage of our algorithm, it is more close to biology reality)</em><br/> | ||
| - | < | + | 2.IPTG molecules can bind to LacR proteins or each monomer of LacR Dimers but cannot bind to LacR dimers whose DNA-binding domains Have been occupied.<br/> |
| + | <em/>(#When there are less LacR proteins, there are less LacR dimers. When there are less LacR dimers, the complex of LacR dimer and DNA is more likely to disassociate.)</em><br/> | ||
| - | + | 3.There are leaky expressions for repressed pLac and pTet promoters.</p> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | <p> | + | <p>In order to test the ability of the system to switch from one state to the other, we conducted a time course simulation. The IPTG are added to the system at time 10000s.<br/> |
| - | + | The GFP expression level changes triggered by the pulse are illustrated by curves with different colors.</p> | |
| - | + | ||
| - | < | + | <img src="https://static.igem.org/mediawiki/2011/6/67/USTC_Software_algo106.jpg" width="512"> |
| - | <p> | + | <p>As you can see from the figure above, at initial time, the LacI protein soon takes control and reaches equilibrium. When IPTG is added to the system, there is a sharp decrease. Since the half life of LacR is short and IPTG is so favorable to bind with LacR, making it inactive (lose the ability to bind tightly to the promoter that helps to express TetR) </p> |
| - | < | + | <h3><a name="RTC two counter (RiboRegulated transcription cascade)" id="RTC two counter (RiboRegulated transcription cascade)"></a>Part2: RTC two counter (RiboRegulated transcription cascade)</h3> |
| - | + | ||
| - | < | + | <p>Recent years saw a emergence of the small RNA application in the gene regulation network. This is a timely way to regulate gene expression compared to other transcription level modulation.</p> |
| - | + | ||
| - | < | + | <P>As a mimic to the electronic digital circuit that can count the pulses or other events (a basic function of most MCU module), synthetic biologist designed ways to count biological events such as the adding of inducers. Here, we adopted the RTC-two counter as an example.</p> |
| - | + | <img src="https://static.igem.org/mediawiki/2011/e/ee/USTC_Software_algo107.jpg" width="604"> | |
| - | <p> | + | <p>The constitutive promoter pl tet 0-1 , which corresponds to j23100 part, drives the T7 RNA polymerase T7RNAP (i2032 part), the RNAP binds to THE T7 promoter PT7 and the GFP will be expressed then.</p> |
| - | + | <P>Between them is the part j01010, which consist of the rbs sequence and the complementary sequence cr ( so the cr section of the mRna transcribed will form a loop with the Rbs section ,thus inhibiting translation)</p> | |
| - | <p> | + | <p>B0014 is the T7 promoter. The RNAP binds to the T7 promoter to drive the expression of the downstream gene (GFP). But still, the translation is inhibited by the stem loop formed by the cr and RBS section of j01010.</p> |
| - | <p> | + | <!--<img src="https://static.igem.org/mediawiki/2011/7/70/USTC_Software_algo108.jpg">--> |
| + | </html> | ||
| + | [[File:rtc2counter_lachesis.jpg|500px]] | ||
| + | <html> | ||
| + | <p>To release the inhibition, the arabinose induced promoter PBAD can drive the expression of | ||
| + | J01008, which corresponds to taRNA. But this promoter can be repressed by c0080, which is the AraC protein dimer. When arabinoses are added to the system, the arabinoses bind to the AraC monomers and prevent them from forming dimers. Thus release the repression to the i01008 promoter.</p> | ||
| - | + | <p>The taRNA is specific to the cr sequence and it bind to the cr sequence, thus open the stem loop, allowing the expression of RNAP and GFP.</p> | |
| - | <p> | + | |
| - | + | <p>The input file to this case is as below:</p> | |
| - | <p> | + | |
| - | < | + | <img src="https://static.igem.org/mediawiki/2011/1/1b/USTC_Software_algo109.jpg"> |
| - | + | <p>The network view generated by our software is as below (this visualization is realized by Junyuan Xie, Fangming Liu, and Chuocheng He. You can drag the nodes to your preferred place.</p> | |
| - | < | + | <img src="https://static.igem.org/mediawiki/2011/9/94/USTC_Software_algo110.jpg" width="644"> |
| - | <p> | + | <p>The list of species generated by the algorithm is listed the table below.</p> |
| - | < | + | <img src="https://static.igem.org/mediawiki/2011/8/88/USTC_Software_algo111.jpg"> |
| - | + | <p>Below is some assumptions made by Liao Chen who worked out this example with his algorithm.<br/> | |
| + | 1.AraC proteins cannot degrade so as to keep forever repression of pBAD before any pulse of arabinose.<br/> | ||
| + | 2.AraC protein can form dimers. But only the dimers, rather than the monomers,can bind to pBAD and repress the expressions of the | ||
| + | downstream genes.<br/> | ||
| + | 3.Arabinose molecules can bind to AraC proteins or each monomer of AraC dimers but cannot bind to AraC dimers whose DNA-binding | ||
| + | Domains have been occupied.<br/> | ||
| + | 4.No leaky expressions is possible for both pBAD and T7 promoter, Which can reduce the leaky GFP expression (noise) before the second.</p> | ||
| - | + | <p>Pulse comes and make the leap of GFP amount more impressive and distinguishable.</p> | |
| - | + | ||
| - | <p> | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | <p>We also attest the validity of our approach using the time course simulation in this case.</p> | ||
| + | <img src="https://static.igem.org/mediawiki/2011/b/b5/USTC_Software_algo112.jpg"> | ||
| + | <p>As you can see from above, two Arabinose pulses are added to the system at time 2500.01s and 2561.01s. The black curve shows the normalized GFP expression of the cells that received both the two pulses. The red line refers to the cells that only receive the first arabinose pulse and the blue line refers to the cells that only receive the second one.</p> | ||
| + | <p>It is clear from the figure, that at the point of the pulse, the GFP level begin to leap to a higher state. Since the inducer is removed from the environment after it generate the pulse, the expression of RNAP will not last long so there is only a small leap if the cell received only one pulse, if with the second, the GFP level will leap markedly as indicated by the blackline.</p> | ||
</div> | </div> | ||
| Line 472: | Line 151: | ||
<div id="content_wrapper_bottom"> </div> | <div id="content_wrapper_bottom"> </div> | ||
| + | |||
<!-- end of content_wrapper --> | <!-- end of content_wrapper --> | ||
| + | |||
| + | |||
| + | |||
<div id="mfooter"> | <div id="mfooter"> | ||
Latest revision as of 16:33, 26 October 2011
 USTC-Software
USTC-Software
MoDeL Demo
Part1: toggle switch.

Toggle switch is characterized by mutual inhibitory network. That is, the protein product of one gene represses the promoter of the other, and vice versa. So with proper parameters, the system will stay at one stable steady state at equilibrium. In other words, one gene's product will be domanant. But with proper inducers (usually IPTG or ATC), the system manage to flop from one state to another, still keeping steady.
We tried an instance of a toggle switch composed of a promoter which can be repressed by laci protein, and is denoted as pLac(r0010). And the other promoter is tet-repressible and be denoted by pTet(r0040). The plac promoter promotes the expression of TetR(c0040), And the LacR(c0012) is initiated by pTet(r0040).
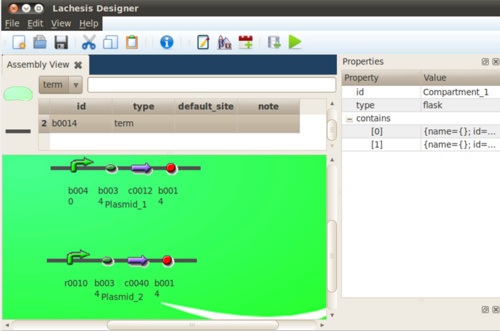
Without inducers, which protein will dominate depend on the binding affinity of the repressor to the promoter. In this case, it's the LacR protein take control at null input of inducers. The only input to the algorithm is a input file like this.

Here are a bit explanations to the input file
As to the seedspecies section:
# Medium iptg : IPTG is contained in the compartment named medium
# Ecoli dna1 d:r0040(tetr1,tetr2)-b0034(rib)-c0012(dna,iptg,dim)-b0014()
#dna1_init
# d:means it's a strand of dna composed of 4 parts.
# The promoters have two binding site(tetr1 and tetr2) for associative repressors.
# c0012(dna,iptg,dim) means the c0012 is a DNA sequence. Iptg means the product #of c0012, which is LacI protein, in terms of dimer, can be binded to iptg which #deactivate it's repression to placi promoter.
#b0014()is the terminator.
#dna1_init means the initial concentration of dna1 is dana1_init which is given in the parameter section of the input file.
Once the input file is provided, both the net and SBML file will be generated within a second. The species "discovered" by the algorithm is illustrated below, totally 18 substances in such a simple network.

A bit explanations are given to the table above.
1.
#nb: non-biobrick part, indicating a constraint of only one part constituting a sequence, making it significantly distinguished from DNA, RNA, Protein sequences.
#d: means the following thing is a sequence of dna with structure information.
# b0014() is a terminator with no binding site.
# the hyphen sign connects the parts on a dna.
#C0012*(dim,dna,iptg) means the c0012 is a dna sequence, and the product of c0012 which is #laci protein, can form dimers, iptg can bind to monomers of laci .
2.
As you can see from species 12 to 15, the advantage of this approach is that it contains information about the structure of the species. For instance, s12 is nb:i0001(laci!1).p:c0012(dim!2,dna,iptg!1).p:c0012(dim!2,dna,iptg)
The ! is used to denote binding , here iptg bind to the laci denoted by !1, and the . is used to separate different molecules of one species. The molecule that comprise the s12 species are nb:i0001(laci!1),p:c0012(dim!2,dna,iptg!1),p:c0012(dim!2,dna,iptg).
The two proteins bind to form dimers through bingding !2.
So now you can imagine the structure of s12 species.
The network generated by our software is as below. User can drag the nodes to the place they want to have a better clarity of the network.

One thing to note is the assumptions made by Chen LIAO:
1.Both LacR protein and TetR protein can form dimers. But only the dimers, rather than the monomers, can bind to the promoter regions
to repress the expressions of the downstream genes.
(#note that this a an advantage of our algorithm, it is more close to biology reality)
2.IPTG molecules can bind to LacR proteins or each monomer of LacR Dimers but cannot bind to LacR dimers whose DNA-binding domains Have been occupied.
(#When there are less LacR proteins, there are less LacR dimers. When there are less LacR dimers, the complex of LacR dimer and DNA is more likely to disassociate.)
3.There are leaky expressions for repressed pLac and pTet promoters.
In order to test the ability of the system to switch from one state to the other, we conducted a time course simulation. The IPTG are added to the system at time 10000s.
The GFP expression level changes triggered by the pulse are illustrated by curves with different colors.

As you can see from the figure above, at initial time, the LacI protein soon takes control and reaches equilibrium. When IPTG is added to the system, there is a sharp decrease. Since the half life of LacR is short and IPTG is so favorable to bind with LacR, making it inactive (lose the ability to bind tightly to the promoter that helps to express TetR)
Part2: RTC two counter (RiboRegulated transcription cascade)
Recent years saw a emergence of the small RNA application in the gene regulation network. This is a timely way to regulate gene expression compared to other transcription level modulation.
As a mimic to the electronic digital circuit that can count the pulses or other events (a basic function of most MCU module), synthetic biologist designed ways to count biological events such as the adding of inducers. Here, we adopted the RTC-two counter as an example.

The constitutive promoter pl tet 0-1 , which corresponds to j23100 part, drives the T7 RNA polymerase T7RNAP (i2032 part), the RNAP binds to THE T7 promoter PT7 and the GFP will be expressed then.
Between them is the part j01010, which consist of the rbs sequence and the complementary sequence cr ( so the cr section of the mRna transcribed will form a loop with the Rbs section ,thus inhibiting translation)
B0014 is the T7 promoter. The RNAP binds to the T7 promoter to drive the expression of the downstream gene (GFP). But still, the translation is inhibited by the stem loop formed by the cr and RBS section of j01010.

To release the inhibition, the arabinose induced promoter PBAD can drive the expression of J01008, which corresponds to taRNA. But this promoter can be repressed by c0080, which is the AraC protein dimer. When arabinoses are added to the system, the arabinoses bind to the AraC monomers and prevent them from forming dimers. Thus release the repression to the i01008 promoter.
The taRNA is specific to the cr sequence and it bind to the cr sequence, thus open the stem loop, allowing the expression of RNAP and GFP.
The input file to this case is as below:

The network view generated by our software is as below (this visualization is realized by Junyuan Xie, Fangming Liu, and Chuocheng He. You can drag the nodes to your preferred place.

The list of species generated by the algorithm is listed the table below.

Below is some assumptions made by Liao Chen who worked out this example with his algorithm.
1.AraC proteins cannot degrade so as to keep forever repression of pBAD before any pulse of arabinose.
2.AraC protein can form dimers. But only the dimers, rather than the monomers,can bind to pBAD and repress the expressions of the
downstream genes.
3.Arabinose molecules can bind to AraC proteins or each monomer of AraC dimers but cannot bind to AraC dimers whose DNA-binding
Domains have been occupied.
4.No leaky expressions is possible for both pBAD and T7 promoter, Which can reduce the leaky GFP expression (noise) before the second.
Pulse comes and make the leap of GFP amount more impressive and distinguishable.
We also attest the validity of our approach using the time course simulation in this case.

As you can see from above, two Arabinose pulses are added to the system at time 2500.01s and 2561.01s. The black curve shows the normalized GFP expression of the cells that received both the two pulses. The red line refers to the cells that only receive the first arabinose pulse and the blue line refers to the cells that only receive the second one.
It is clear from the figure, that at the point of the pulse, the GFP level begin to leap to a higher state. Since the inducer is removed from the environment after it generate the pulse, the expression of RNAP will not last long so there is only a small leap if the cell received only one pulse, if with the second, the GFP level will leap markedly as indicated by the blackline.
 "
"
