Team:HKU-Hong Kong/Modelling
From 2011.igem.org
| (8 intermediate revisions not shown) | |||
| Line 6: | Line 6: | ||
|- | |- | ||
|style="width:900px;"| | |style="width:900px;"| | ||
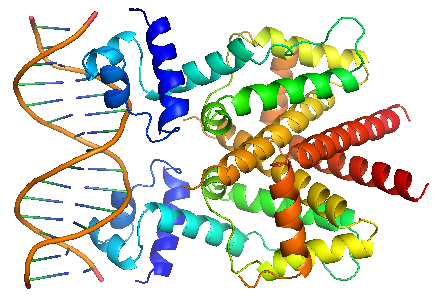
| - | TetR regulates the most abundant resistance mechanism against the antibiotic tetracycline in gram-negative bacteria. It consists of 208 residues, being folded into a 10 α – helices. α1 – α3 forms the DNA-binding domain, within which α2 – α3 constitute the classical helix-turn-helix motifs. α5 – α10 forms the binding domain of the tetracycline – magnesium complex, [MgTc]+, α5, α8 and α10 are helices of the rigid scaffold, within which α8 and α10 form the central part of the regulatory domain with the dyad related α8’ and α10’. α4 acts as the hydrophobic center of the DNA binding domain, and links the DNA-binding domain to the regulatory domain. | + | TetR regulates the most abundant resistance mechanism against the antibiotic tetracycline in gram-negative bacteria. It consists of 208 residues, being folded into a 10 α – helices. α1 – α3 forms the DNA-binding domain, within which α2 – α3 constitute the classical helix-turn-helix motifs. α5 – α10 forms the binding domain of the tetracycline – magnesium complex, [MgTc]+, α5, α8 and α10 are helices of the rigid scaffold, within which α8 and α10 form the central part of the regulatory domain with the dyad related α8’ and α10’. α4 acts as the hydrophobic center of the DNA binding domain, and links the DNA-binding domain to the regulatory domain. |
<div ALIGN=CENTER> | <div ALIGN=CENTER> | ||
{| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | {| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| Line 12: | Line 12: | ||
|[[Image:TetR.png|250px]] | |[[Image:TetR.png|250px]] | ||
|- | |- | ||
| - | |TetR | + | |TetR binding with TetO |
|} | |} | ||
</div> | </div> | ||
|} | |} | ||
| - | {|style="width:900px;"| | + | {| style="width:900px;background:#000000;text-align:justify;font-family: georgia, helvetica, arial, sans-serif;color:#ffffff;margin-top:25px;" cellspacing="20" |
| + | |style="width:900px;"| | ||
TetR binds with tetO under normal circumstances. In the DNA-bound complex, the two-fold symmetry of TetR is maintained. Each HTH-motif binds to the corresponding major groove of the palindromic tetO, while the minor groove is not recognized. All but the central 3 pairs of the 15-mer operator fragment are engaged in the binding. | TetR binds with tetO under normal circumstances. In the DNA-bound complex, the two-fold symmetry of TetR is maintained. Each HTH-motif binds to the corresponding major groove of the palindromic tetO, while the minor groove is not recognized. All but the central 3 pairs of the 15-mer operator fragment are engaged in the binding. | ||
| Line 28: | Line 29: | ||
|} | |} | ||
</div> | </div> | ||
| + | |} | ||
| + | |||
| + | {| style="width:900px;background:#000000;text-align:justify;font-family: georgia, helvetica, arial, sans-serif;color:#ffffff;margin-top:25px;" cellspacing="20" | ||
| + | |style="width:900px;"| | ||
| + | TetR binds with tetO under normal circumstances. In the DNA-bound complex, the two-fold symmetry of TetR is maintained. Each HTH-motif binds to the corresponding major groove of the palindromic tetO, while the minor groove is not recognized. All but the central 3 pairs of the 15-mer operator fragment are engaged in the binding. | ||
| + | In the presence of tetracycline – magnesium complex, [MgTc]+, residue His100 and Thr103 of helix α6 are displaced, causing the shifting of helix α6 towards the C terminal. Both C and N terminal of α4 shift because of the movement of α6 and the formation of hydrogen bond between α4 and [MgTc]+. α3 is displaced as a consequence, which releases the bonding between tetR and tetO.<br/> | ||
| + | |} | ||
| + | |||
| + | {| style="width:900px;background:#000000;text-align:justify;font-family: georgia, helvetica, arial, sans-serif;color:#ffffff;margin-top:25px;" cellspacing="20" | ||
| + | |style="width:900px; font-size:1.5em;color:#CC0000;"|DNA-binding protein H-NS | ||
| + | |- | ||
| + | |style="width:900px;"| | ||
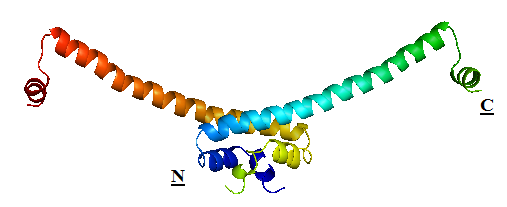
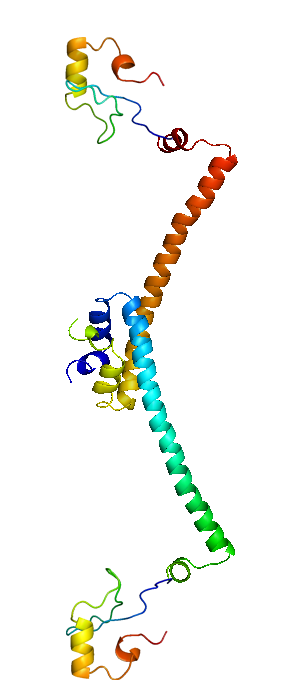
| + | H-NS proteins are a type of histone-like protein found in abundance in Gram-negative bacteria (such as E.coli). Phylogenetic studies have shown there are some varieties of H-NS proteins in different bacteria species. Nonetheless, H-NS are involved in regulating transcription by repression, and the structuring of the nucleoid. The regulation of H-NS expression is complex. Its expression is likely to be influenced by the external environment as well. However, it is also capable of autoregulating its own transcriptions. | ||
| + | H-NS regulates gene expression negatively; whilst a complete understanding of its mechanism is yet to be established due to limitations of current lab techniques, research has identified a number of common themes: first, H-NS proteins acts on (non-specifically) intrinsically curved DNA sites near the promoter of the gene to be repressed. Intrinsically curved DNA sites near the promoter enable upstream and downstream DNA to be brought in close proximity after the binding of the RNA polymerase on the promoter. Bound H-NS on these regions interacts to form a nucleoprotein complex that traps RNA polymerase, thus effectively stopping transcription. The trapping of the RNA polymerase on the promoter means it need not be recruited from the cytoplasm again once repression is relieved, hence allowing a rapid response to changes in environmental conditions. | ||
| + | <div ALIGN=CENTER> | ||
| + | {| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:HNS_(1-90).png|250px]] | ||
| + | |- | ||
| + | |HNS 1 - 90 | ||
| + | |} | ||
| + | </div> | ||
| + | <div ALIGN=CENTER> | ||
| + | {| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:HNS_(1-137).png|250px]] | ||
| + | |- | ||
| + | |HNS 1 - 137 | ||
| + | |} | ||
| + | </div> | ||
| + | The structure of H-NS protein is vital to its gene repressing activities. The H-NS protein contains 136 amino acids, giving rise to three distinct regions: the N-terminal domain (a.a. 1-64), the linker region (a.a. 65-89) and the C-terminal domain (a.a. 90-137). The linker region is flexible and connects the two domains together. An isolated N-terminal domain is capable to undergo dimerisation on its own: indeed, a 46-residue segment of the domain has been identified to be the minimal dimerisation domain. On the other hand, the C-terminal domain contains a highly conserved motif (a.a. 108-116) that allows the H-NS protein to bind to nucleic acid. Dimerisation of H-NS on binding to the nucleic acid provides the basic unit for subsequent oligomerisation. According to one model, a.a. 46-90 is the central region for oligomerisation activity. The H-NS dimer nucleates along the DNA from its initial binding site to create a nucleoprotein complex that effectively ‘zipper’ stretches of DNA and represses gene transcription.<br/> | ||
| + | |} | ||
| + | |||
| + | {| style="width:900px;background:#000000;text-align:justify;font-family: georgia, helvetica, arial, sans-serif;color:#ffffff;margin-top:25px;" cellspacing="20" | ||
| + | |style="width:900px; font-size:1.5em;color:#CC0000;"|Gene regulation by HNS-tetR | ||
| + | |- | ||
| + | |style="width:900px;"| | ||
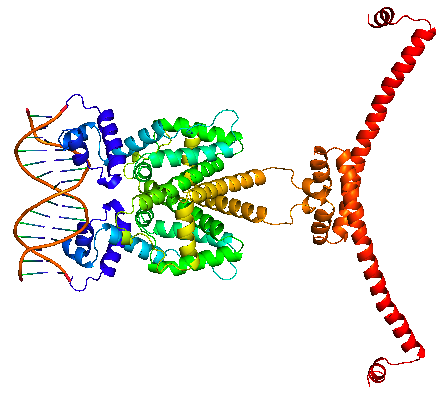
| + | The HNS-tetR fusion protein is arranged in and NC-NC manner, binding with its operator DNA. Through the subsequent oligomerization of HNS protein along the DNA, DNA is stretched and gene expression is suppressed. | ||
| + | <div ALIGN=CENTER> | ||
| + | {| style="width:254px;background:#99EE63;text-align:center;font-family: georgia, helvetica, arial, sans-serif;color:#000000;margin- top:5px;padding: 2px;" cellspacing="5"; | ||
| + | |- | ||
| + | |[[Image:TetR-HNS-TetO.png|450px]] | ||
| + | |- | ||
| + | |HNS-TetR binding with tetO | ||
| + | |} | ||
| + | </div> | ||
| + | <br/> | ||
|} | |} | ||
Latest revision as of 12:15, 5 October 2011
| Modelling |
| The tetracycline inducible Tet repressor-operator system |
|
TetR regulates the most abundant resistance mechanism against the antibiotic tetracycline in gram-negative bacteria. It consists of 208 residues, being folded into a 10 α – helices. α1 – α3 forms the DNA-binding domain, within which α2 – α3 constitute the classical helix-turn-helix motifs. α5 – α10 forms the binding domain of the tetracycline – magnesium complex, [MgTc]+, α5, α8 and α10 are helices of the rigid scaffold, within which α8 and α10 form the central part of the regulatory domain with the dyad related α8’ and α10’. α4 acts as the hydrophobic center of the DNA binding domain, and links the DNA-binding domain to the regulatory domain. |
|
TetR binds with tetO under normal circumstances. In the DNA-bound complex, the two-fold symmetry of TetR is maintained. Each HTH-motif binds to the corresponding major groove of the palindromic tetO, while the minor groove is not recognized. All but the central 3 pairs of the 15-mer operator fragment are engaged in the binding. In the presence of tetracycline – magnesium complex, [MgTc]+, residue His100 and Thr103 of helix α6 are displaced, causing the shifting of helix α6 towards the C terminal. Both C and N terminal of α4 shift because of the movement of α6 and the formation of hydrogen bond between α4 and [MgTc]+. α3 is displaced as a consequence, which releases the bonding between tetR and tetO. |
|
TetR binds with tetO under normal circumstances. In the DNA-bound complex, the two-fold symmetry of TetR is maintained. Each HTH-motif binds to the corresponding major groove of the palindromic tetO, while the minor groove is not recognized. All but the central 3 pairs of the 15-mer operator fragment are engaged in the binding.
In the presence of tetracycline – magnesium complex, [MgTc]+, residue His100 and Thr103 of helix α6 are displaced, causing the shifting of helix α6 towards the C terminal. Both C and N terminal of α4 shift because of the movement of α6 and the formation of hydrogen bond between α4 and [MgTc]+. α3 is displaced as a consequence, which releases the bonding between tetR and tetO. |
| DNA-binding protein H-NS |
|
H-NS proteins are a type of histone-like protein found in abundance in Gram-negative bacteria (such as E.coli). Phylogenetic studies have shown there are some varieties of H-NS proteins in different bacteria species. Nonetheless, H-NS are involved in regulating transcription by repression, and the structuring of the nucleoid. The regulation of H-NS expression is complex. Its expression is likely to be influenced by the external environment as well. However, it is also capable of autoregulating its own transcriptions. H-NS regulates gene expression negatively; whilst a complete understanding of its mechanism is yet to be established due to limitations of current lab techniques, research has identified a number of common themes: first, H-NS proteins acts on (non-specifically) intrinsically curved DNA sites near the promoter of the gene to be repressed. Intrinsically curved DNA sites near the promoter enable upstream and downstream DNA to be brought in close proximity after the binding of the RNA polymerase on the promoter. Bound H-NS on these regions interacts to form a nucleoprotein complex that traps RNA polymerase, thus effectively stopping transcription. The trapping of the RNA polymerase on the promoter means it need not be recruited from the cytoplasm again once repression is relieved, hence allowing a rapid response to changes in environmental conditions. The structure of H-NS protein is vital to its gene repressing activities. The H-NS protein contains 136 amino acids, giving rise to three distinct regions: the N-terminal domain (a.a. 1-64), the linker region (a.a. 65-89) and the C-terminal domain (a.a. 90-137). The linker region is flexible and connects the two domains together. An isolated N-terminal domain is capable to undergo dimerisation on its own: indeed, a 46-residue segment of the domain has been identified to be the minimal dimerisation domain. On the other hand, the C-terminal domain contains a highly conserved motif (a.a. 108-116) that allows the H-NS protein to bind to nucleic acid. Dimerisation of H-NS on binding to the nucleic acid provides the basic unit for subsequent oligomerisation. According to one model, a.a. 46-90 is the central region for oligomerisation activity. The H-NS dimer nucleates along the DNA from its initial binding site to create a nucleoprotein complex that effectively ‘zipper’ stretches of DNA and represses gene transcription. |
| Gene regulation by HNS-tetR |
|
The HNS-tetR fusion protein is arranged in and NC-NC manner, binding with its operator DNA. Through the subsequent oligomerization of HNS protein along the DNA, DNA is stretched and gene expression is suppressed.
|
 "
"