Team:ITESM Mexico/Assembly
From 2011.igem.org
(Difference between revisions)
| (5 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
<html><h1 class="myowntitle">Assembly</h1> </html> | <html><h1 class="myowntitle">Assembly</h1> </html> | ||
| + | ------ | ||
| Line 8: | Line 9: | ||
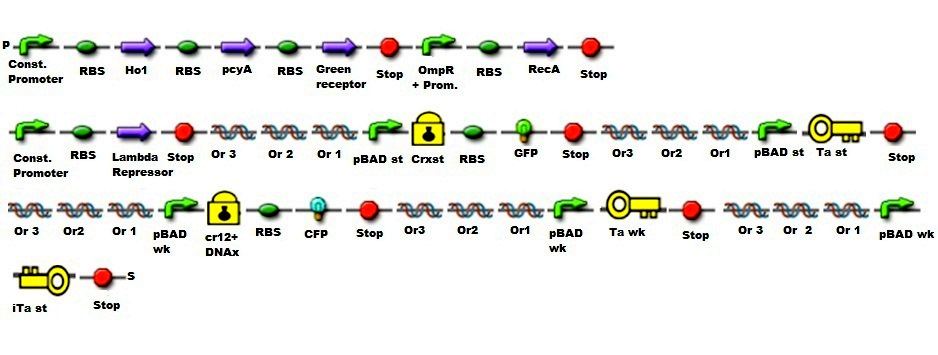
| - | ::::::::[[File:Part1.jpg| | + | ::::::::[[File:Part1.jpg|400px]] Assembly for the photoreceptor |
| - | ::::::::[[File:Part2.jpg| | + | ::::::::[[File:Part2.jpg|400px]] Assembly for high and low concentration |
| Line 19: | Line 20: | ||
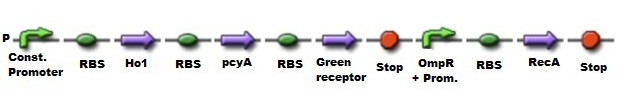
[[File:Const.Promoter.jpg|100px|caption]] | [[File:Const.Promoter.jpg|100px|caption]] | ||
<html></td><td></html> | <html></td><td></html> | ||
| - | '''BBa_J23109:''' Constitutive promoter family. | + | '''[http://partsregistry.org/Part:BBa_J23109 BBa_J23109:]''' Constitutive promoter family. |
Parts J23100 through J23119 are a family of constitutive promoter parts isolated from a small combinatorial library. J23119 is the "consensus" promoter sequence and the strongest member of the family. All parts except J23119 are present in plasmid J61002. Part J23119 is present in pSB1A2. This places the RFP downstream of the promoter. Reported activities of the promoters are given as the relative fluorescence of these plasmids in strain TG1 grown in LB media to saturation. See part J61002 for details on their use. These promoter parts can be used to tune the expression level of constitutively expressed parts. The NheI and AvrII restriction sites present within these promoter parts make them a scaffold for further modification. | Parts J23100 through J23119 are a family of constitutive promoter parts isolated from a small combinatorial library. J23119 is the "consensus" promoter sequence and the strongest member of the family. All parts except J23119 are present in plasmid J61002. Part J23119 is present in pSB1A2. This places the RFP downstream of the promoter. Reported activities of the promoters are given as the relative fluorescence of these plasmids in strain TG1 grown in LB media to saturation. See part J61002 for details on their use. These promoter parts can be used to tune the expression level of constitutively expressed parts. The NheI and AvrII restriction sites present within these promoter parts make them a scaffold for further modification. | ||
Ampicillin resistant. | Ampicillin resistant. | ||
| Line 27: | Line 28: | ||
[[File:RBS.jpg|100px|caption]] | [[File:RBS.jpg|100px|caption]] | ||
<html></td><td></html> | <html></td><td></html> | ||
| - | '''BBa_B0034:''' RBS based on Elowitz repressilator. | + | '''[http://partsregistry.org/wiki/index.php/Part:BBa_B0034 BBa_B0034:]''' RBS based on Elowitz repressilator. |
Ampicillin resistant. | Ampicillin resistant. | ||
<html></td></tr></html> | <html></td></tr></html> | ||
| Line 34: | Line 35: | ||
[[File:HO1.jpg|100px|caption]] | [[File:HO1.jpg|100px|caption]] | ||
<html></td><td></html> | <html></td><td></html> | ||
| - | '''BBa_I15008:''' One of two requisite genes required for the biosynthesis of phycocyanobilin from heme. | + | '''[http://partsregistry.org/Part:BBa_I15008 BBa_I15008:]''' One of two requisite genes required for the biosynthesis of phycocyanobilin from heme. |
Usage and Biology | Usage and Biology | ||
Ho1 oxidizes the heme group using a ferredoxin cofactor, generating biliverdin IXalpha and representing the first of two steps in phycocyanobilin (PCB) biosynthesis. PCB associates with Cph8, creating a light responsive protein complex. Functions in tandem with BBa_I15009 for PCB biosynthesis. PCB then associates withPart:BBa_I15010, a light responsive Cph8/EnvZ fusion protein. | Ho1 oxidizes the heme group using a ferredoxin cofactor, generating biliverdin IXalpha and representing the first of two steps in phycocyanobilin (PCB) biosynthesis. PCB associates with Cph8, creating a light responsive protein complex. Functions in tandem with BBa_I15009 for PCB biosynthesis. PCB then associates withPart:BBa_I15010, a light responsive Cph8/EnvZ fusion protein. | ||
| Line 43: | Line 44: | ||
[[File:pcyA.jpg|100px|caption]] | [[File:pcyA.jpg|100px|caption]] | ||
<html></td><td></html> | <html></td><td></html> | ||
| - | '''BBa_I15009:''' Phycocyanobilin:ferredoxin oxidoreductase (PcyA) from synechocystis | + | '''[http://partsregistry.org/Part:BBa_I15009 BBa_I15009:]''' Phycocyanobilin:ferredoxin oxidoreductase (PcyA) from synechocystis |
Second of two required phycocyanobilin biosynthetic genes. | Second of two required phycocyanobilin biosynthetic genes. | ||
Usage and Biology: Converts biliverdin IXalpha (BV)(produced by Part:BBa_I15008) to phycocyanobilin (PCB), the immediate precursor of cyanobacterial phytochromes. Associates with Part:BBa_I15010 to transduce a light signal into a genetic response. Kanamycin resistant. | Usage and Biology: Converts biliverdin IXalpha (BV)(produced by Part:BBa_I15008) to phycocyanobilin (PCB), the immediate precursor of cyanobacterial phytochromes. Associates with Part:BBa_I15010 to transduce a light signal into a genetic response. Kanamycin resistant. | ||
| Line 51: | Line 52: | ||
[[File:Green_receptor.jpg|100px|caption]] | [[File:Green_receptor.jpg|100px|caption]] | ||
<html></td><td></html> | <html></td><td></html> | ||
| - | '''BBa_K225000:''' Green light receptor. | + | '''[http://partsregistry.org/Part:BBa_K225000 BBa_K225000:]''' Green light receptor. |
Fusion protein of light responsive domains of CcaS from Synechocystis sp. PCC 6803 and histidine kinase EnvZ. For the use as a green light activated actuator in E. coli. | Fusion protein of light responsive domains of CcaS from Synechocystis sp. PCC 6803 and histidine kinase EnvZ. For the use as a green light activated actuator in E. coli. | ||
Usage and Biology: GlrN is the fusion protein of light responsive domains of CcaS and EnvZ designed by changing the histidine kinase region of CcaS to EnvZ domain of cph8 (BBa_I15010). | Usage and Biology: GlrN is the fusion protein of light responsive domains of CcaS and EnvZ designed by changing the histidine kinase region of CcaS to EnvZ domain of cph8 (BBa_I15010). | ||
| - | + | *Requires phycocyanobilin biosynthetic genes (BBa_I15008 and BBa_I15009) for chromophore formation. | |
| - | + | *Must be used in E. coli deficient in natural EnvZ. | |
Expecting function (without experimental proof): Exposure to green light (535 nm) phosphorylate EnvZ histidin kinase domain. The activated EnvZ phosphorylates endogenous OmpR, a transcription factor which activates transcription from the OmpR positive promoter (BBa_R0083). | Expecting function (without experimental proof): Exposure to green light (535 nm) phosphorylate EnvZ histidin kinase domain. The activated EnvZ phosphorylates endogenous OmpR, a transcription factor which activates transcription from the OmpR positive promoter (BBa_R0083). | ||
| Line 64: | Line 65: | ||
[[File:Stop.jpg|100px|caption]] | [[File:Stop.jpg|100px|caption]] | ||
<html></td><td></html> | <html></td><td></html> | ||
| - | '''BBa_B0015:''' Double terminator consisting of BBa_B0010 and BBa_B0012. | + | '''[http://partsregistry.org/Part:BBa_B0015 BBa_B0015:]''' Double terminator consisting of BBa_B0010 and BBa_B0012. |
Ampicillin and Kanamycin resistant. | Ampicillin and Kanamycin resistant. | ||
<html></td></tr></html> | <html></td></tr></html> | ||
| Line 71: | Line 72: | ||
[[File:OmpR+Prom.jpg|100px|caption]] | [[File:OmpR+Prom.jpg|100px|caption]] | ||
<html></td><td></html> | <html></td><td></html> | ||
| - | '''BBa_R0083:''' Promoter (OmpR, positive) | + | '''[http://partsregistry.org/wiki/index.php?title=Part:BBa_R0083 BBa_R0083:]''' Promoter (OmpR, positive) |
Positively regulated, OmpR-controlled promoter. This promoter is derived from the upstream region of ompC. Phosphorylated OmpR binds to the operator site and activates transcription. This is a truncated version of BBa_R0082. | Positively regulated, OmpR-controlled promoter. This promoter is derived from the upstream region of ompC. Phosphorylated OmpR binds to the operator site and activates transcription. This is a truncated version of BBa_R0082. | ||
Usage and Biology: This promoter contains upstream regions of the ompC porin gene. The regulation of ompC is determined by the EnvZ-OmpR osmosensing machinery. EnvZ phosphorylates OmpR to OmpR-P. At high osmolarity, EnvZ is more active, creating more OmpR-P. OmpR-P then binds to the low-affinity OmpR operator sites upstream of ompC. | Usage and Biology: This promoter contains upstream regions of the ompC porin gene. The regulation of ompC is determined by the EnvZ-OmpR osmosensing machinery. EnvZ phosphorylates OmpR to OmpR-P. At high osmolarity, EnvZ is more active, creating more OmpR-P. OmpR-P then binds to the low-affinity OmpR operator sites upstream of ompC. | ||
| Line 88: | Line 89: | ||
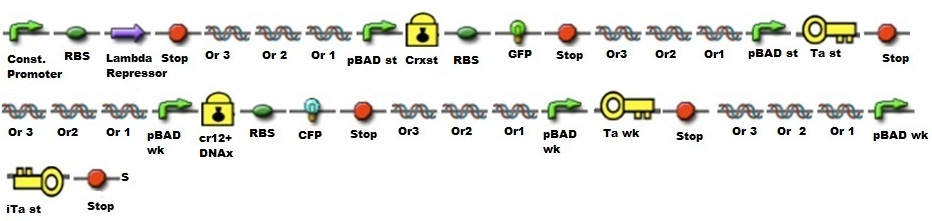
[[File:LambdaRepresor.jpg|100px|caption]] | [[File:LambdaRepresor.jpg|100px|caption]] | ||
<html></td><td></html> | <html></td><td></html> | ||
| - | '''BBa_C0051:''' cI repressor from E. coli phage lambda (+LVA) | + | '''[http://partsregistry.org/Part:BBa_C00513 BBa_C0051:]''' cI repressor from E. coli phage lambda (+LVA) |
Coding region for the cI repressor based on cI repressor from bacteriophage lambda modified with an LVA tail for rapid degradation of the protein. cI repressor binds to the cI regulator (BBa_R0051). | Coding region for the cI repressor based on cI repressor from bacteriophage lambda modified with an LVA tail for rapid degradation of the protein. cI repressor binds to the cI regulator (BBa_R0051). | ||
Ampicillin resistant. | Ampicillin resistant. | ||
| Line 96: | Line 97: | ||
[[File:Or3.jpg|100px|caption]] | [[File:Or3.jpg|100px|caption]] | ||
<html></td><td></html> | <html></td><td></html> | ||
| - | '''BBa_K079043:''' Lambda OR3 - operator library member | + | '''[http://partsregistry.org/Part:BBa_K079043 BBa_K079043:]''' Lambda OR3 - operator library member |
Lambda Genetic switch is a complex and deeply characterized system. | Lambda Genetic switch is a complex and deeply characterized system. | ||
Ampicillin resistant. | Ampicillin resistant. | ||
| Line 104: | Line 105: | ||
[[File:Or2.jpg|100px|caption]] | [[File:Or2.jpg|100px|caption]] | ||
<html></td><td></html> | <html></td><td></html> | ||
| - | '''BBa_K079042:''' Lambda OR2 - operator library member. Lambda Genetic switch is a complex and deeply characterized system. | + | '''[http://partsregistry.org/Part:BBa_K079042 BBa_K079042:]''' Lambda OR2 - operator library member. Lambda Genetic switch is a complex and deeply characterized system. |
Ampicillin resistant. | Ampicillin resistant. | ||
<html></td></tr></html> | <html></td></tr></html> | ||
| Line 111: | Line 112: | ||
[[File:Or1.jpg|100px|caption]] | [[File:Or1.jpg|100px|caption]] | ||
<html></td><td></html> | <html></td><td></html> | ||
| - | '''BBa_K079041:''' Lambda OR1 - operator library member. Lambda Genetic switch is a complex and deeply characterized system. | + | '''[http://partsregistry.org/Part:BBa_K079041 BBa_K079041:]''' Lambda OR1 - operator library member. Lambda Genetic switch is a complex and deeply characterized system. |
Ampicillin resistant. | Ampicillin resistant. | ||
<html></td></tr></html> | <html></td></tr></html> | ||
| Line 118: | Line 119: | ||
[[File:pBADst.jpg|100px|caption]] | [[File:pBADst.jpg|100px|caption]] | ||
<html></td><td></html> | <html></td><td></html> | ||
| - | ''' | + | '''[http://partsregistry.org/Part:BBa_K206000 BBa_K206000:]''' pBAD strong: Inducible promoter that responds to low arabinose concentrations. Click on the part's number for description. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<html></td></tr></html> | <html></td></tr></html> | ||
| Line 145: | Line 132: | ||
[[File:GFP.jpg|100px|caption]] | [[File:GFP.jpg|100px|caption]] | ||
<html></td><td></html> | <html></td><td></html> | ||
| - | '''BBa_K145015:''' GFP with LVA tag | + | '''[http://partsregistry.org/Part:BBa_K145015 BBa_K145015:]''' GFP with LVA tag |
Green Fluorescent Protein with LVA tag for rapid degradation. This part was thoroughly tested by the 2008 KULeuven team. This team showed that this GFP mutant is degraded at least 35x faster than GFP (mut3b) and that it has a half life of about 74 minutes. | Green Fluorescent Protein with LVA tag for rapid degradation. This part was thoroughly tested by the 2008 KULeuven team. This team showed that this GFP mutant is degraded at least 35x faster than GFP (mut3b) and that it has a half life of about 74 minutes. | ||
Ampicillin resistant. | Ampicillin resistant. | ||
| Line 160: | Line 147: | ||
[[File:pBADwk.jpg|100px|caption]] | [[File:pBADwk.jpg|100px|caption]] | ||
<html></td><td></html> | <html></td><td></html> | ||
| - | '''BBa_K206001:''' Name: pBAD weak | + | '''[http://partsregistry.org/Part:BBa_K206001 BBa_K206001:]''' Name: pBAD weak |
Input: L-arabinose | Input: L-arabinose | ||
| Line 184: | Line 171: | ||
[[File:CFP.jpg|100px|caption]] | [[File:CFP.jpg|100px|caption]] | ||
<html></td><td></html> | <html></td><td></html> | ||
| - | '''BBa_E0022:''' Enhanced cyan fluorescent protein derived from A. victoria GFP | + | '''[http://partsregistry.org/wiki/index.php?title=Part:BBa_E0022 BBa_E0022:]''' Enhanced cyan fluorescent protein derived from A. victoria GFP |
Cyan fluorescent protein (ECFP) reporter coding sequence without the Ribosome Binding Site. Modified with an LVA tail for rapid degradation of the protein and faster fall time for the emission. | Cyan fluorescent protein (ECFP) reporter coding sequence without the Ribosome Binding Site. Modified with an LVA tail for rapid degradation of the protein and faster fall time for the emission. | ||
Ampicillin resistant. | Ampicillin resistant. | ||
Latest revision as of 23:42, 14 October 2011
Assembly
 "
"