Team:USC/Project
From 2011.igem.org
(Difference between revisions)
(→Genetic Arrangement of CRISPR/Cas System) |
(→BL21 (DE3) strain w/ tetO::GFP, CRISPR-GFP, cas3, casABCDE12) |
||
| (70 intermediate revisions not shown) | |||
| Line 47: | Line 47: | ||
===Genetic Arrangement of CRISPR/Cas System=== | ===Genetic Arrangement of CRISPR/Cas System=== | ||
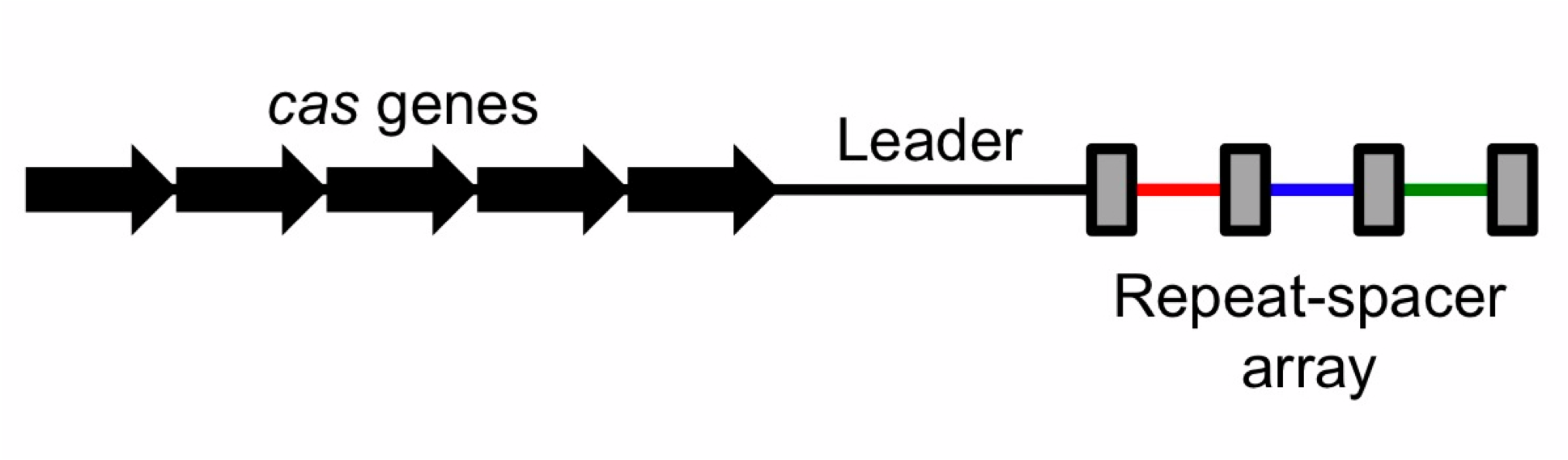
| - | To the right is a schematic diagram of how the CRISPR/Cas system is arranged genetically.[[File:CRISPRschematic.png|300px|thumb| | + | To the right is a schematic diagram of how the CRISPR/Cas system is arranged genetically.[[File:CRISPRschematic.png|300px|thumb|center|Diagram of CRISPR/Cas system. The repeats are displayed as rectangles. Spacers are colored.]] Upstream of the CRISPR locus are a series of cas genes arranged sequentially in the following order: |
*cas3 | *cas3 | ||
| - | *casABCDE12 | + | *casABCDE12 (short for casA, casB, casC...) |
| - | [[File:n-CRISPR.png|600px|thumb|center]] | + | What follows is the promoter of the CRIPSR locus, which has been sequenced to be A-T nucleotide rich. The CRISPR array of repeats and spacers completes the system. |
| - | [[File:CRISPRsetup.png|400px|thumb|center]] | + | |
| + | ===Synthetic Version of CRISPR=== | ||
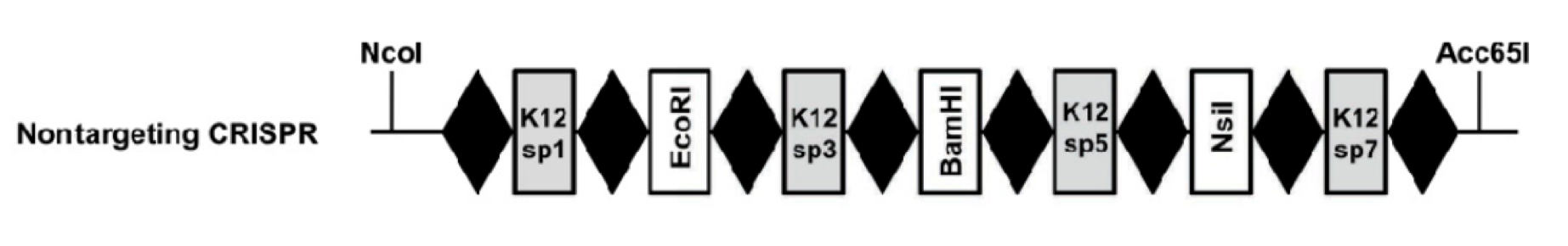
| + | The idea of our project comes from utilizing the ''E. coli'' bacteria's natural way of conferring resistance to foreign DNA and plasmid and see if it can be engineered such that the CRISPR/Cas system can target specifically any piece of DNA by means of programming the system with a spacer of any DNA sequence. Such a system has been engineered by the Laboratory of Microbiology in the University of Wageningen and is displayed below: | ||
| + | [[File:n-CRISPR.png|600px|thumb|center|K12 Inspired CRISPR cassette with alternating K12-targeting spacers and restriction sites]] | ||
| + | This part, BBa_K512002, is an IPTG-inducible CRISPR array genetically engineered with alternating spacers that target the K12 genome and contain restriction sites. The restriction enzyme sites are designed so that a spacer of any DNA sequence can be inserted into this CRISPR system. | ||
| + | |||
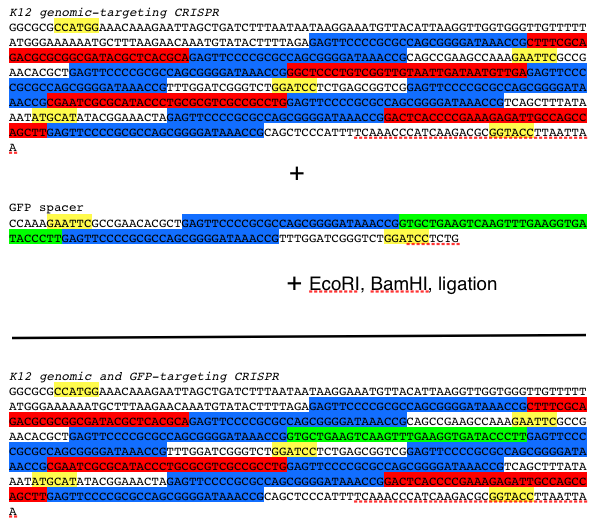
| + | In our particular project, we designed our synthetic CRISPR to target a DNA sequence coding for GFP. To do so, we took the synthetic CRISPR and GFP spacer and digested with two compatible restriction enzyme so that the spacer can be properly ligated into our synthetic CRISPR. The protocol is displayed below: | ||
| + | [[File:CRISPRsetup.png|400px|thumb|center|Protocol for constructing CRISPR-GFP: yellow=restriction enzyme site, blue=repeat, red=K12 targeting spacer, green=GFP-targeting spacer]] | ||
<h1 style="font-family:Verdana;font-weight:500;">Results</h1> | <h1 style="font-family:Verdana;font-weight:500;">Results</h1> | ||
| + | Once the CRISPR-GFP plasmid has been constructed, we decided to test the system in K12 and B ''E. coli'': | ||
| + | |||
| + | ===CRISPR/Cas System in K12 E. coli=== | ||
| + | To test the CRISPR/Cas system in K12, we designed the following strains: | ||
| + | *HT115 (K12) strain w/ tetO::GFP (AmpR) | ||
| + | *HT115 (K12) strain w/ tetO::GFP & CRISPR-GFP (AmpR & CmR) | ||
| + | *HT115 (K12) strain w/ CRISPR-GFP (CmR) | ||
| + | We tested each system by creating three batches for each strain so we can monitor the growth of the bacteria over time: | ||
| + | *One strain + antibiotics - IPTG | ||
| + | *Same strain + antibiotics + IPTG | ||
| + | *Same strain + antibiotics + 1 hr. before inducing system with IPTG | ||
| + | |||
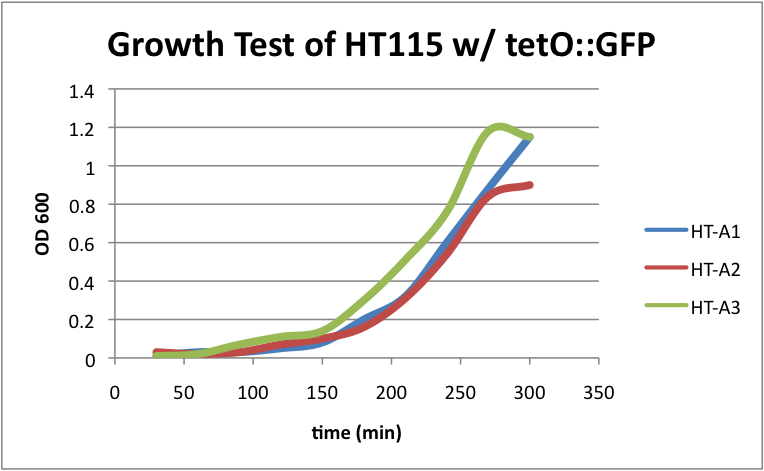
| + | ====HT115 w/ tetO::GFP==== | ||
| + | Regarding the first strain (HT115 w/ tetO::GFP), we used this test as our control. | ||
| + | [[File:HT115grow1.png|400px|thumb|center|HT-A1=(-IPTG), HT-A2=(+IPTG), HT-A3=(1hr.+IPTG)]] | ||
| + | The three batches of the strain grew at approximately the same rates. | ||
| + | |||
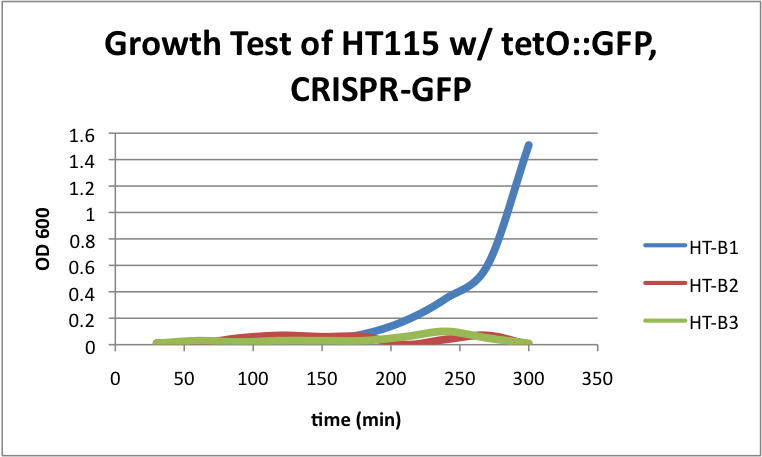
| + | ====HT115 w/ tetO::GFP & CRISPR-GFP==== | ||
| + | |||
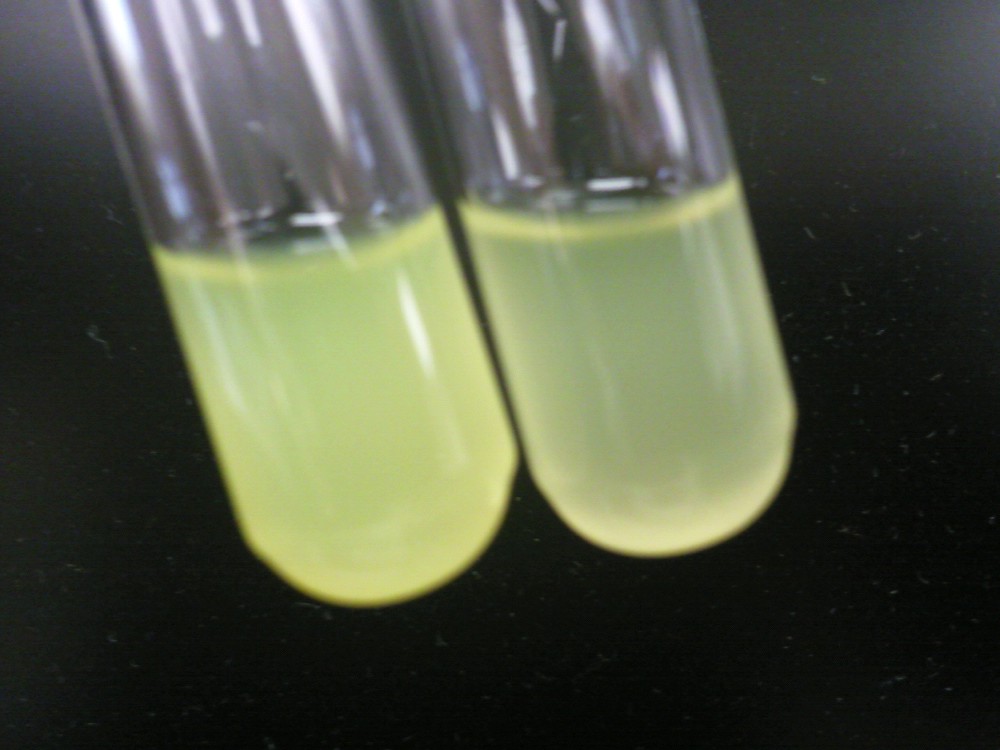
| + | [[File:HT115g1.jpeg|200px|thumb|center|Left: HT-B1 overnight; Right: HT-B2 overnight]] | ||
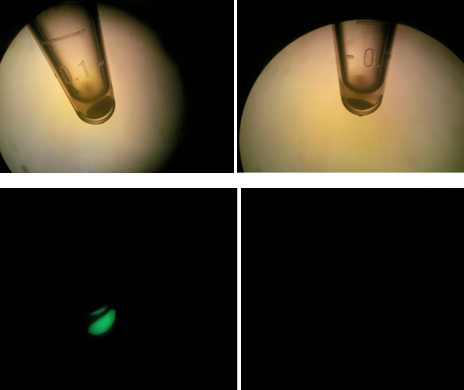
| + | [[File:HT115g2.png|200px|thumb|center|Upper & lower left: HT-B1; Upper & lower right: HT-B2]] | ||
| + | [[File:HT115grow2.png|400px|thumb|center|HT-B1=(-IPTG), HT-B2=(+IPTG), HT-B3=(1hr.+IPTG)]] | ||
| + | |||
| + | With the addition of CRISPR-GFP, there is a severe drop off in growth rate for the batches induced with IPTG. This indicates that CRISPR-GFP system is destroying the tetO::GFP plasmid that also contains the antibiotic resistance gene for ampicillin. Since the bacteria are growing in cultures containing ampicillin, the activated CRISPR renders the bacteria incapable of surviving in the presence of ampicllin. As a result, they are no longer able to survive. | ||
| + | |||
| + | Above, the picture diagrams show our test for tetO::GFP. After the growth test, we clearly observed that the batch HT-B1 was more dense than that of HT-B2. More importantly, under a fluorescence scope, we also observed significant GFP expression from HT-B1. However, no GFP expression could be observed from HT-B2. This indicates that HT-B1 still harbors the tetO::GFP plasmid, whereas HT-B2 does not. | ||
| + | |||
| + | ====HT115 w/ CRISPR-GFP==== | ||
| + | [[File:HT115grow3.png|400px|thumb|center|HT-D1=(-IPTG), HT-D2=(+IPTG), HT-D3=(1hr.+IPTG)]] | ||
| + | This test is used to confirm that CRISPR can also target and destroy genomic DNA from the K12 family. Note that this CRISPR system also contains spacers that match the genomic DNA of K12 bacteria. Although this data introduces a bias to the growth test of strain HT115 with tetO::GFP & CRISPR-GFP, we are encouraged by the observation that the batches of bacterial strain HT115 with CRISPR-GFP induced by IPTG grew slightly more before dying from the presence of ampicillin. | ||
| + | |||
| + | ===CRISPR/cas System in B E. coli=== | ||
| + | |||
| + | To test the CRISPR/Cas system in B, we designed the following strains: | ||
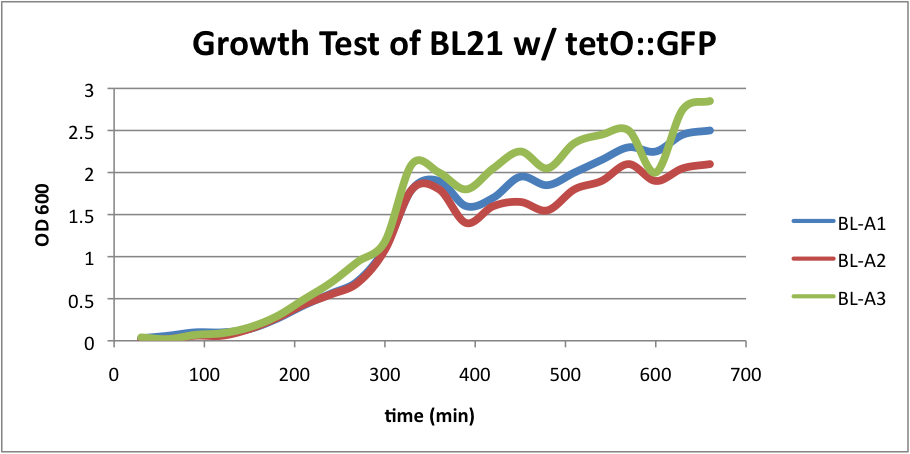
| + | *BL21 (DE3) strain w/ tetO::GFP (AmpR) | ||
| + | *BL21 (DE3) strain w/ tetO::GFP & CRISPR-GFP (AmpR & CmR) | ||
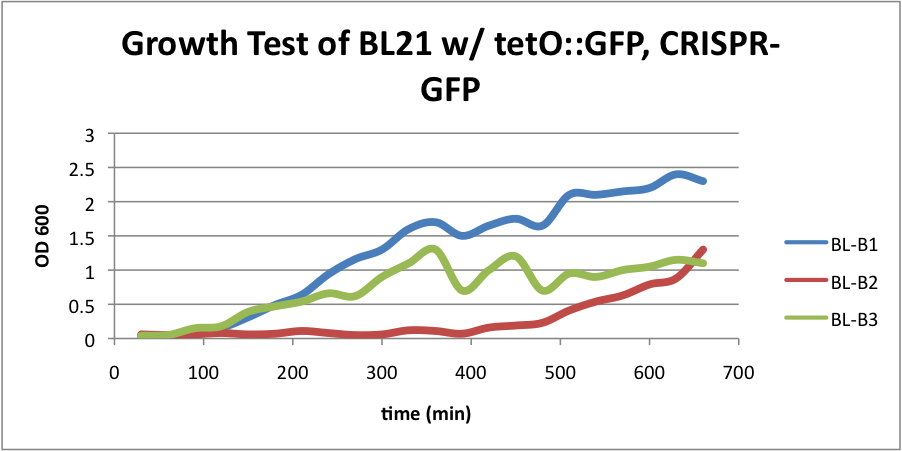
| + | *BL21 (DE3) strain w/ tetO::GFP, CRISPR-GFP, cas3, casABCDE (AmpR, CmR, KanR, StrepR) | ||
| + | *BL21 (DE3) strain w/ tetO::GFP, CRISPR-GFP, cas3, casABCDE12 (AmpR, CmR, KanR, StrepR) | ||
| + | |||
| + | We tested each system by creating three batches for each strain so we can monitor the growth of the bacteria over time: | ||
| + | *One strain + antibiotics - IPTG | ||
| + | *Same strain + antibiotics + IPTG | ||
| + | *Same strain + antibiotics + 2 hr. before inducing system with IPTG | ||
| + | |||
| + | ====BL21 (DE3) strain w/ tetO::GFP==== | ||
| + | Regarding the first strain (BL21 w/ tetO::GFP), we used this test as our control. | ||
| + | [[File:BL21grow1.png|400px|thumb|center|BL-A1=(-IPTG), BL-A2=(+IPTG), BL-A3=(2hr.+IPTG)]] | ||
| + | The three batches of the strain grew at approximately the same rates. | ||
| + | |||
| + | ====BL21 (DE3) strain w/ tetO::GFP & CRISPR-GFP==== | ||
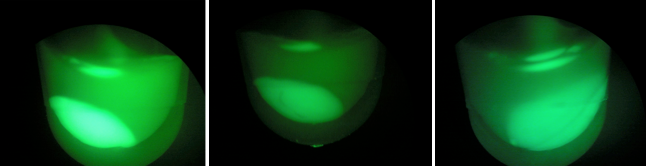
| + | [[File:B1B2B3.jpeg|200px|thumb|center|BL-B1=(-IPTG), BL-B2=(+IPTG), BL-B3=(2hr.+IPTG)]] | ||
| + | [[File:gfpb1b2b3.jpeg|200px|thumb|center|Left:BL-B1=(-IPTG), Middle:BL-B2=(+IPTG), Right:BL-B3=(2hr.+IPTG)]] | ||
| + | [[File:BL21grow2.png|400px|thumb|center|BL-B1=(-IPTG), BL-B2=(+IPTG), BL-B3=(2hr.+IPTG)]] | ||
| + | |||
| + | The growth test for BL21 (DE3) strain with tetO::GFP & CRISPR-GFP shows interesting results. Although we expected growth rates to be similar to our control strain, we instead observed lowered growth rates in batches induced with IPTG. Although studies have shown that CRIPSR interference against viruses in ''E. coli'' is governed by cas3, casABCDE, and CRISPR, this may indicate that CRISPR interference against plasmids in vivo may be governed by a slightly different cocktail of genes. This is a test we look forward to repeating. | ||
| + | |||
| + | Above, the picture diagrams display our test for GFP expression. After the growth test, we observed that BL-B1 was the most dense of the batches. In addition, it displayed the brightest GFP expression. These observations substantiate the possibility the CRISPR-GFP works under a mechanism that may require different genes or no cas genes. | ||
| + | |||
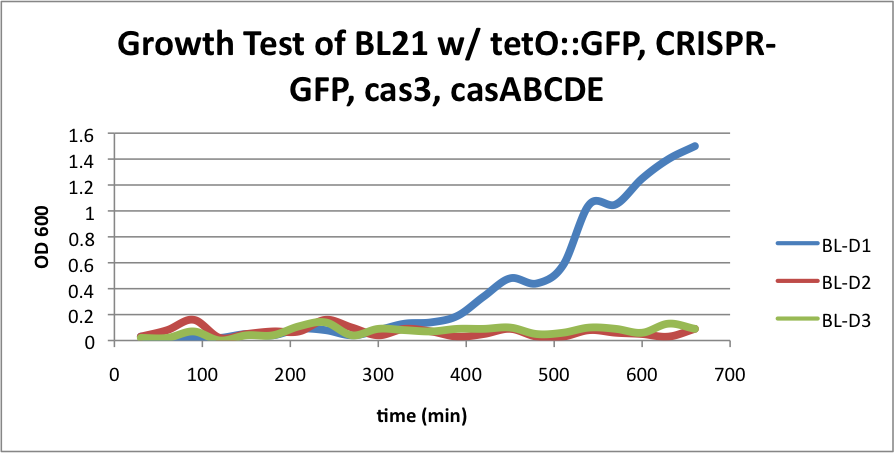
| + | ====BL21 (DE3) strain w/ tetO::GFP, CRISPR-GFP, cas3, casABCDE==== | ||
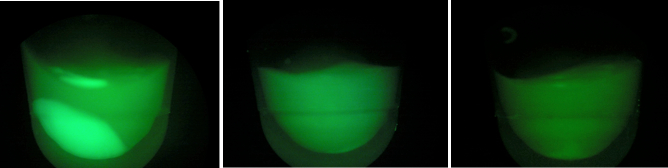
| + | [[File:D1D2D3.jpeg|200px|thumb|center|BL-D1=(-IPTG), BL-D2=(+IPTG), BL-D3=(2hr.+IPTG)]] | ||
| + | [[File:gfpd1d2d3.jpeg|200px|thumb|center|Left:BL-D1=(-IPTG), Middle:BL-D2=(+IPTG), Right:BL-D3=(2hr.+IPTG)]] | ||
| + | [[File:BL21grow3.png|400px|thumb|center|BL-B1=(-IPTG), BL-B2=(+IPTG), BL-B3=(2hr.+IPTG)]] | ||
| - | + | Compared to the growth test of strain BL21 with tetO::GFP & CRISPR-GFP, we have a stronger CRISPR interference response as shown in the growth test curve. | |
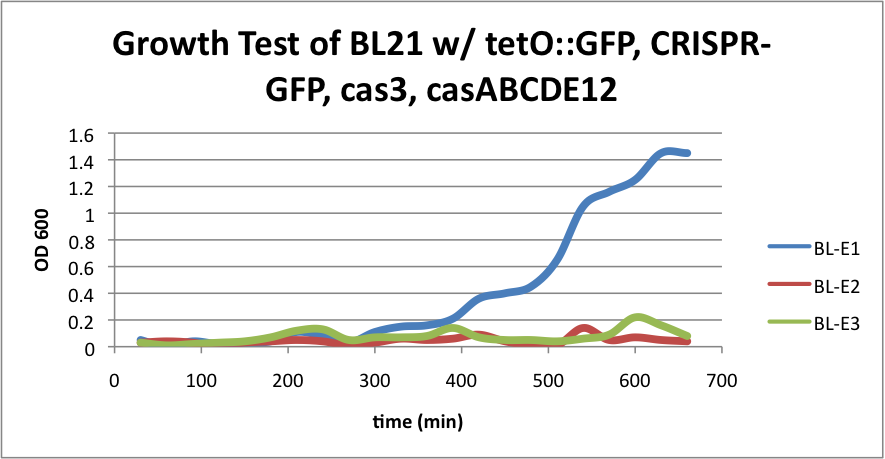
| - | + | ====BL21 (DE3) strain w/ tetO::GFP, CRISPR-GFP, cas3, casABCDE12==== | |
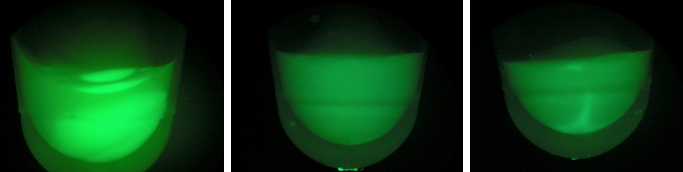
| - | [[File: | + | [[File:E1E2E3.jpeg|200px|thumb|center|BL-E1=(-IPTG), BL-E2=(+IPTG), BL-E3=(2hr.+IPTG)]] |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | [[File:gfpe1e2e3.jpeg|200px|thumb|center|Left:BL-E1=(-IPTG), Middle:BL-E2=(+IPTG), Right:BL-E3=(2hr.+IPTG)]] | |
| + | [[File:BL21grow4.png|400px|thumb|center|BL-E1=(-IPTG), BL-E2=(+IPTG), BL-E3=(2hr.+IPTG)]] | ||
| - | + | Compared to the growth test of strain BL21 with tetO::GFP & CRISPR-GFP, we have a stronger CRISPR interference response as shown in the growth test curve. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | =Future Directions= | |
| - | + | CRISPR is an effective method for plasmid curing, as seen by our own results. Although further research much be conducted to validate its ability to destroy all types of plasmids in bacterial cells, the future of CRISPR is positive. CRISPR, for example, has the potential to combat plasmids that carry antibiotic-resistant genes to other bacteria, hence reversing the evolution of antibiotic-resistance in bacteria. Moreover, we can prevent harmful phages from destroying certain bacteria by programming CRISPR to target foreign DNA. | |
| - | + | ||
| + | There are numerous benefits of CRISPR, from medical to environmental. However, CRISPR’s potential will not be fully realized until further research is accomplished. We must still figure out the effects combinations of spacers and CRISPR cassettes have on plasmids curing. We must also discover ways to practically implement these systems. Although there is much left to learn, CRISPR is definitely a promising area of synthetic biology. | ||
 "
"