Team:UTP-Panama/Project
From 2011.igem.org
(→History: The original idea: July 15) |
(→Project Description) |
||
| (64 intermediate revisions not shown) | |||
| Line 42: | Line 42: | ||
|- | |- | ||
|style="background-image: url(https://static.igem.org/mediawiki/2011/1/13/Fndo1.png); background-color: #FFFFFF" width="75px" valign="top"| | |style="background-image: url(https://static.igem.org/mediawiki/2011/1/13/Fndo1.png); background-color: #FFFFFF" width="75px" valign="top"| | ||
| + | <div> | ||
| + | <br>[[Image:Logoigempeginav1.png|150px|center|link=https://2011.igem.org/Main_Page]] | ||
| + | </div> | ||
<br><center> | <br><center> | ||
[[Team:UTP-Panama/Home | <font face="verdana" style="color:#663366"> '''Home''' </font>]] <br><br> | [[Team:UTP-Panama/Home | <font face="verdana" style="color:#663366"> '''Home''' </font>]] <br><br> | ||
| Line 47: | Line 50: | ||
[[Team:UTP-Panama/Project | <font face="verdana" style="color:#663366"> '''The Project''' </font>]] <br><br> | [[Team:UTP-Panama/Project | <font face="verdana" style="color:#663366"> '''The Project''' </font>]] <br><br> | ||
[[Team:UTP-Panama/Parts | <font face="verdana" style="color:#663366"> '''Parts''' </font>]] <br><br> | [[Team:UTP-Panama/Parts | <font face="verdana" style="color:#663366"> '''Parts''' </font>]] <br><br> | ||
| - | [[Team:UTP-Panama/Modelling| <font face="verdana" style="color:#663366"> '''Project Design''' </font>]] <br><br> | + | [[Team:UTP-Panama/Modelling| <font face="verdana" style="color:#663366"> '''Project & Experiments Design''' </font>]] <br><br> |
[[Team:UTP-Panama/Human Practice| <font face="verdana" style="color:#663366"> '''Human Practice''' </font>]] <br><br> | [[Team:UTP-Panama/Human Practice| <font face="verdana" style="color:#663366"> '''Human Practice''' </font>]] <br><br> | ||
[[Team:UTP-Panama/Notebook | <font face="verdana" style="color:#663366"> '''Notebook''' </font>]] <br><br> | [[Team:UTP-Panama/Notebook | <font face="verdana" style="color:#663366"> '''Notebook''' </font>]] <br><br> | ||
| Line 53: | Line 56: | ||
[[Team:UTP-Panama/Data Page| <font face="verdana" style="color:#663366"> '''Data Page''' </font>]] <br><br> | [[Team:UTP-Panama/Data Page| <font face="verdana" style="color:#663366"> '''Data Page''' </font>]] <br><br> | ||
[[Team:UTP-Panama/Sponsors| <font face="verdana" style="color:#663366"> '''Sponsors''' </font>]] <br><br> | [[Team:UTP-Panama/Sponsors| <font face="verdana" style="color:#663366"> '''Sponsors''' </font>]] <br><br> | ||
| + | [[File:Renbo-move.gif|Center]] | ||
|width="880" valign="top" style="padding: 10px; border: 5px solid #660000; color: #000; background-color: #white" | | |width="880" valign="top" style="padding: 10px; border: 5px solid #660000; color: #000; background-color: #white" | | ||
| Line 64: | Line 68: | ||
[[Image:Image-Size.gif|center]] | [[Image:Image-Size.gif|center]] | ||
| - | |||
| + | == INTRODUCTION == | ||
| + | <div align="justify">In the engineering field, robust sensors are needed, and they must be able to work under different conditions and ranges. | ||
| + | In past iGEM editions, there were attempts to get more and more organisms, such as machines & sensors, to be functional and resistant into multiple stress conditions. | ||
| - | + | The UTP-Panama team in its vision to develop engineering tools from biotechnology, seeks to create and improve more robust sensors to environmental and climatic conditions. | |
| - | + | In this sense, one of the biggest challenges is to give microorganisms the ability to continue working under temperature changes, since they loose capacity at low temperatures (D'amico, et al. 2006). This problem has been attacked by different teams, like the UNAM-Cinvestav 2010, using the CspA promoter as a better promoter for low temperatures in combination with an antifreeze protein (AFP) [1]. Our Team, the UTP-Panama seeks to extend the range of sensors from the use of the AOX enzyme (Alternative oxidase), which primarily has been designed to be expressed in response to a Cold-shock, raising the temperature as we explain next. | |
| - | + | The biosensor chosen for the improvements is the Nitrate Biosensor (PyeaR - composite GFP) developed by BCCS-Bristol Team 2010, which is a sensor that measures levels of nitrogen (nutrients) on the ground, (important parameter for agriculture)</div> | |
| + | == Project Description== | ||
| + | <div align="justify"> | ||
| + | The original BioBrick that senses the levels of nitrates in soil developed by Bristol iGEM Team 2010 [2], was designed to operate at room temperatures, especially for 37 ° C using the PyeaR BioBricks promoter [3]. | ||
| + | Our team seeks to extend the range of optimal functioning of the sensor developed by Bristol at temperatures below | ||
| + | 20 ° C, as many of the crops in highlands (coffee and fruits), are in this range. To solve this problem we found that the use of the AOX enzyme (Alternative oxidase) gene responsible for thermogenesis in the sacred lotus [4], mainly used in Synthetic Biology to generate an increase in periplasmic temperature, produced an interesting increase in the growth and downsizing bacterial densities in temperature [5]. This part uses a HybB promoter [6] for "cold shock". | ||
| + | We, the UTP-Panama iGEM Team developed a new "device" by Combining the two parts, the AOX periplasmatic heat generator (BioBrick BBa_K410000) and the Nitrates biosensor (BioBrick BBa_K381001), to create that we call The Renbo, to ensure that our meters Nutrient operate at low temperatures. The Renbo is the first of a family of Improved Biosensor that the UTP-Panama Team plans to create. | ||
| - | + | </div> | |
| + | ===Future Develops=== | ||
| + | <div align="justify"> | ||
| + | The next step in our project is to interchange the HybB promoter by the CspA promoter (BBa_K328001), these will produce an anti freeze protein that prevents ice crystals formation in the cell, by applying these we expect E.coli to survive and also perceive an increase on the genetic expression of our Escherichia Coli and still be able to sense the nitrate and nitrites on even lower temperatures than 15ºC (limit temperature of RENBO). | ||
| + | <br> | ||
| + | </div> | ||
| + | ====The original BrainStorms ideas==== | ||
| - | = | + | <html> |
| + | To see the Original Project Description June-July 2011 click | ||
| + | <A href="https://2011.igem.org/Team:UTP-Panama/Project_firstidea"> here</A>. | ||
| + | </html> | ||
| + | ==Our Project Design== | ||
| + | <div align="justify"> | ||
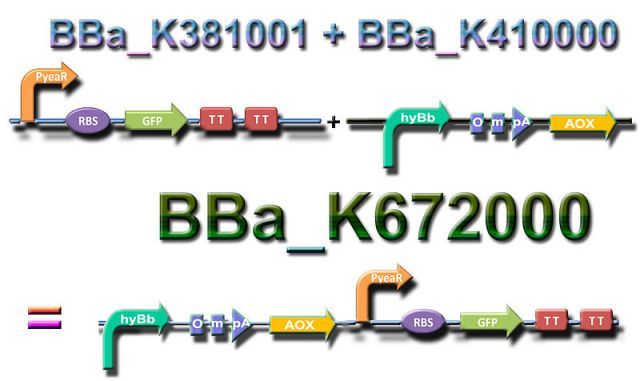
| + | The Bristol BBa_k381001 encodes an expression of GFP in response of Nitrate or Nitrites that expresses fluorescent signals upon nutrient detection, its principal application is that allows farmers to quantify soil nutrient content. | ||
| + | The BBa_k410000 is a fusion of the HydB cold shock promoter, the OmpA a signal peptide, and AOX 1 . AOXa is a alternative oxidase found in Sacred Lotus. This is going to produce a increase of heat above ambient temperature as follow: | ||
| + | · From 20 °C to 30 ° C the HydB promoter will express.<br> | ||
| + | · Approximately at 37° the promoter will stop his expression. | ||
| + | By combining BBa_k38100 and BBa_k410000 we expected our part , the BBa_K672000 , to be able to sense nitrate and nitrites even after experiencing a cold shock at 20º C, making our BB employable in other places where the temperatures can be as low as 15º C. | ||
| + | </div> | ||
| - | + | [[File:BBCOMBINATIONreordered.jpg|center]] | |
| + | == REFERENCES== | ||
| + | [1]https://2010.igem.org/Team:Mexico-UNAM-CINVESTAV/Project/Our_project<br> | ||
| + | [2]http://partsregistry.org/Part:BBa_K381001<br> | ||
| + | [3]http://partsregistry.org/Part:BBa_K216005<br> | ||
| + | [4]Grant, N et al. “Two Cys or Not Two Cys? That is the Question; Alternative Oxidase in the Thermogenic Plant sacred Lotus.” Plant Physiology 150 (2009): 987-995.<br> | ||
| + | [5]Cell Density and Growth Rate Monitoring Tables. (available at http://partsregistry.org/Part:BBa_K410000).<br> | ||
| + | [6]http://partsregistry.org/Part:BBa_J45503 | ||
| - | == | + | ==Aknowledgements== |
| + | To: Dr Nigel Savery | ||
| + | University of Bristol | ||
| + | To help us to understand the AgrEcoli Experiments and Project. | ||
Latest revision as of 05:33, 27 October 2011
|
Home |
THERMOGENIC RESPONSE NUTRIENT BIOSENSOR (THE RENBO)
INTRODUCTIONIn the engineering field, robust sensors are needed, and they must be able to work under different conditions and ranges.
In past iGEM editions, there were attempts to get more and more organisms, such as machines & sensors, to be functional and resistant into multiple stress conditions. The UTP-Panama team in its vision to develop engineering tools from biotechnology, seeks to create and improve more robust sensors to environmental and climatic conditions. In this sense, one of the biggest challenges is to give microorganisms the ability to continue working under temperature changes, since they loose capacity at low temperatures (D'amico, et al. 2006). This problem has been attacked by different teams, like the UNAM-Cinvestav 2010, using the CspA promoter as a better promoter for low temperatures in combination with an antifreeze protein (AFP) [1]. Our Team, the UTP-Panama seeks to extend the range of sensors from the use of the AOX enzyme (Alternative oxidase), which primarily has been designed to be expressed in response to a Cold-shock, raising the temperature as we explain next. The biosensor chosen for the improvements is the Nitrate Biosensor (PyeaR - composite GFP) developed by BCCS-Bristol Team 2010, which is a sensor that measures levels of nitrogen (nutrients) on the ground, (important parameter for agriculture)Project DescriptionThe original BioBrick that senses the levels of nitrates in soil developed by Bristol iGEM Team 2010 [2], was designed to operate at room temperatures, especially for 37 ° C using the PyeaR BioBricks promoter [3]. Our team seeks to extend the range of optimal functioning of the sensor developed by Bristol at temperatures below 20 ° C, as many of the crops in highlands (coffee and fruits), are in this range. To solve this problem we found that the use of the AOX enzyme (Alternative oxidase) gene responsible for thermogenesis in the sacred lotus [4], mainly used in Synthetic Biology to generate an increase in periplasmic temperature, produced an interesting increase in the growth and downsizing bacterial densities in temperature [5]. This part uses a HybB promoter [6] for "cold shock". We, the UTP-Panama iGEM Team developed a new "device" by Combining the two parts, the AOX periplasmatic heat generator (BioBrick BBa_K410000) and the Nitrates biosensor (BioBrick BBa_K381001), to create that we call The Renbo, to ensure that our meters Nutrient operate at low temperatures. The Renbo is the first of a family of Improved Biosensor that the UTP-Panama Team plans to create.
Future DevelopsThe next step in our project is to interchange the HybB promoter by the CspA promoter (BBa_K328001), these will produce an anti freeze protein that prevents ice crystals formation in the cell, by applying these we expect E.coli to survive and also perceive an increase on the genetic expression of our Escherichia Coli and still be able to sense the nitrate and nitrites on even lower temperatures than 15ºC (limit temperature of RENBO).
The original BrainStorms ideasTo see the Original Project Description June-July 2011 click here. Our Project DesignThe Bristol BBa_k381001 encodes an expression of GFP in response of Nitrate or Nitrites that expresses fluorescent signals upon nutrient detection, its principal application is that allows farmers to quantify soil nutrient content. The BBa_k410000 is a fusion of the HydB cold shock promoter, the OmpA a signal peptide, and AOX 1 . AOXa is a alternative oxidase found in Sacred Lotus. This is going to produce a increase of heat above ambient temperature as follow: · From 20 °C to 30 ° C the HydB promoter will express. By combining BBa_k38100 and BBa_k410000 we expected our part , the BBa_K672000 , to be able to sense nitrate and nitrites even after experiencing a cold shock at 20º C, making our BB employable in other places where the temperatures can be as low as 15º C.
REFERENCES[1]https://2010.igem.org/Team:Mexico-UNAM-CINVESTAV/Project/Our_project AknowledgementsTo: Dr Nigel Savery University of Bristol To help us to understand the AgrEcoli Experiments and Project. |
 "
"