Team:UNAM-Genomics Mexico/Notebook/hydaopt
From 2011.igem.org
| (3 intermediate revisions not shown) | |||
| Line 14: | Line 14: | ||
| - | '''''CAI optimization and 5'UTR ΔGibbs optimization control''''' | + | '''''CAI optimization and 5'UTR ΔGibbs optimization control'''''. |
| + | |||
| + | |||
| + | ''See also our [[Team:UNAM-Genomics_Mexico/Optimization|optimization protocol]].'' | ||
| Line 43: | Line 46: | ||
| - | '''pBBR1MCS-5''' derived plasmid. This plasmid is a modification of the pBBR1MCS-5 plasmid, which is a broad-host-range (bhr) vector designed to assist genetic anaylsis in prokaryotes, specifically for Gram- bacteria that are naturally CmR (chloramphenicol resistant). It is used to transfer genes from E. coli to R. etli by first transforming E. coli S17 competent cells with this vector containing an RFP reporter and later performing a conjugation between the transformed E. coli cells and R. etli CFN42. | + | '''pBBR1MCS-5''' derived plasmid. This plasmid is a modification of the pBBR1MCS-5 plasmid, which is a broad-host-range (bhr) vector designed to assist genetic anaylsis in prokaryotes, specifically for Gram- bacteria that are naturally CmR (chloramphenicol resistant). It is used to transfer genes from ''E. coli'' to ''R. etli'' by first transforming ''E. coli'' S17 competent cells with this vector containing an RFP reporter and later performing a conjugation between the transformed ''E. coli'' cells and ''R. etli'' CFN42. |
| Line 55: | Line 58: | ||
| - | '''HydA optimized CDS'''. This sequence was made by the bioinformatic tools we developed that change all the codons in the sequence by the most used codons by the | + | '''HydA optimized CDS'''. This sequence was made by the bioinformatic tools we developed that change all the codons in the sequence by the most used codons by the constitutively expressed genes in ''Rhizobium etli'' CFN42. It is 1749 nucleotide long. |
| - | Two constructions are built, each composed of the vector pBBR1MCS-5, the part Bba_J04500, the part Bba_B0015 and differing in the coding region each one | + | Two constructions are built, each composed of the vector pBBR1MCS-5, the part Bba_J04500, the part Bba_B0015 and differing in the coding region each one possesses, either the wild-type or the optimized hydrogenase I, HydA gene. |
| Line 66: | Line 69: | ||
===Measurement=== | ===Measurement=== | ||
| - | Once assembled, the plasmids should be transformed to S17 E. coli cells, and latter conjugated to Rhizobium etli CFN42. Once in its final host, both R. etli will be put to test their growing performances. As the one carrying the plasmids that has the optimized version of the hydrogenase, HydA, has the appropiate codons that allow the cell to overexpress the gene without effectively affecting the pool of tRNAs existing within the bacteria, it is expected to be the one with the better growth curves; compared to the one that carries the wild-type version of HydA. We've chose just to test the optimization control with just one gene, and specially with the hydrogenase gene, as we are not expecting to produce molecular hydrogen and because we do not want to affect the growth of the bacteria by using R. etli metabolites with the system. The ferredoxin and the PFOR were not added to this system. Even so, we cannot completely asure that there will not be hydrogen production, as other endogenous redox potential donnors might interact with the hydrogenase. | + | Once assembled, the plasmids should be transformed to S17 ''E. coli'' cells, and latter conjugated to ''Rhizobium etli'' CFN42. Once in its final host, both ''R. etli'' will be put to test their growing performances. As the one carrying the plasmids that has the optimized version of the hydrogenase, HydA, has the appropiate codons that allow the cell to overexpress the gene without effectively affecting the pool of tRNAs existing within the bacteria, it is expected to be the one with the better growth curves; compared to the one that carries the wild-type version of HydA. We've chose just to test the optimization control with just one gene, and specially with the hydrogenase gene, as we are not expecting to produce molecular hydrogen and because we do not want to affect the growth of the bacteria by using ''R. etli'' metabolites with the system. The ferredoxin and the PFOR were not added to this system. Even so, we cannot completely asure that there will not be hydrogen production, as other endogenous redox potential donnors might interact with the hydrogenase. |
===Justification=== | ===Justification=== | ||
| - | Our main objective with the project Hydrobium etli is the production of molecular hydrogen that is profitable to use it as an energy carrier. In order to achieve this, we must ensure that the system we've introduced into Rhizobium etli functions at its best conditions, firing on all cyllinders; but at the same time without affecting the bacterium host. This last implication is of utmost importance, as the bacteria R. etli is symbiont of the plant Phaseolus vulgaris. The health of the bacteria is necessary for the very plant survival. | + | Our main objective with the project Hydrobium etli is the production of molecular hydrogen that is profitable to use it as an energy carrier. In order to achieve this, we must ensure that the system we've introduced into ''Rhizobium etli'' functions at its best conditions, firing on all cyllinders; but at the same time without affecting the bacterium host. This last implication is of utmost importance, as the bacteria ''R. etli'' is symbiont of the plant ''Phaseolus vulgaris''. The health of the bacteria is necessary for the very plant survival. |
| Line 78: | Line 81: | ||
*Paul M.Sharpl and Wen-Hsiung Li. The codon adaptation index - a measure of directional synonymous codon usage bias, and its potential application. Nucleic Acids Research. 1987. | *Paul M.Sharpl and Wen-Hsiung Li. The codon adaptation index - a measure of directional synonymous codon usage bias, and its potential application. Nucleic Acids Research. 1987. | ||
| - | *Grzegorz Kudla, Andrew W. Murray, David Tollervey, Joshua B. Plotkin. Coding-Sequence Determinants of Gene Expression in Escherichia coli. Sicence. 2009. | + | *Grzegorz Kudla, Andrew W. Murray, David Tollervey, Joshua B. Plotkin. Coding-Sequence Determinants of Gene Expression in ''Escherichia coli''. Sicence. 2009. |
| - | *Mark Welch Sridhar Govindarajan, Jon E. Ness, Alan Villalobos, Austin Gurney, Jeremy Minshull, Claes Gustafsson. Design Parameters to Control Synthetic Gene Expression in Escherichia coli. PLoS one, 2009 | + | *Mark Welch Sridhar Govindarajan, Jon E. Ness, Alan Villalobos, Austin Gurney, Jeremy Minshull, Claes Gustafsson. Design Parameters to Control Synthetic Gene Expression in ''Escherichia coli''. PLoS one, 2009 |
*Sivan Navon, Yitzhak Pilpel. The role of codon selection in regulation of translation efficiency deduced from synthetic libraries. Genome Biology. 2011 | *Sivan Navon, Yitzhak Pilpel. The role of codon selection in regulation of translation efficiency deduced from synthetic libraries. Genome Biology. 2011 | ||
| - | *Kuchenreuther JM, Grady-Smith CS, Bingham AS, George SJ, Cramer SP, et al. (2010) High-Yield Expression of Heterologous [FeFe] Hydrogenases in Escherichia coli. PLoS ONE 5(11): e15491 | + | *Kuchenreuther JM, Grady-Smith CS, Bingham AS, George SJ, Cramer SP, et al. (2010) High-Yield Expression of Heterologous [FeFe] Hydrogenases in ''Escherichia coli''. PLoS ONE 5(11): e15491 |
*Agapakis et al.: Insulation of a synthetic hydrogen metabolism circuit in bacteria. Journal of Biological Engineering 2010 4:3. | *Agapakis et al.: Insulation of a synthetic hydrogen metabolism circuit in bacteria. Journal of Biological Engineering 2010 4:3. | ||
Latest revision as of 04:30, 29 September 2011
Optimization of HydA
Contents |
CAI optimization and 5'UTR ΔGibbs optimization control.
See also our optimization protocol.
General Overview
Overview
One of the main aims in Synthetic Biology is the design and construction of new biological functions and systems not found in nature. One can accomplish this by assembling a new genetic system from scratch by adding elements (genes) from different species and introducing it into a new host. Each organism has evolve in a manner that permits the differential usage of codons to a specific set of genes; that is, constitutively expressed genes have their own usage of tRNAs, and so on. If an exogenous sequence is to be expressed in a host, and this happens to use some rare codons that have been reserved to other endogenous sequences, the host might result compromised in its ability to properly translate this endogenous sequences and so, its growth might get affected. It is of our concern not to affect the host of our genetic circuit, Rhizobium etli, that's way we've developed bioinformatic tools that allow us to modify the genes that form our system in a manner that they have the codons used by the constitutively expressed genes of R. etli; by doing this, we expect not to harm the bacteria by expressing our system.
We also modified the 5'UTR and a small portion of the coding region in the amino terminus that minimizes the probability of forming secondary structures that hinder the ribosome from binding the transcript and start the translation.
As all of our codifying region of our system has been optimized for CAI and for 5'UTR ΔGibbs, we expect that by comparing wild-type sequences against our optimized, to observe a growth difference between the Rhizobium etli carrying either of these variants.
Control Components
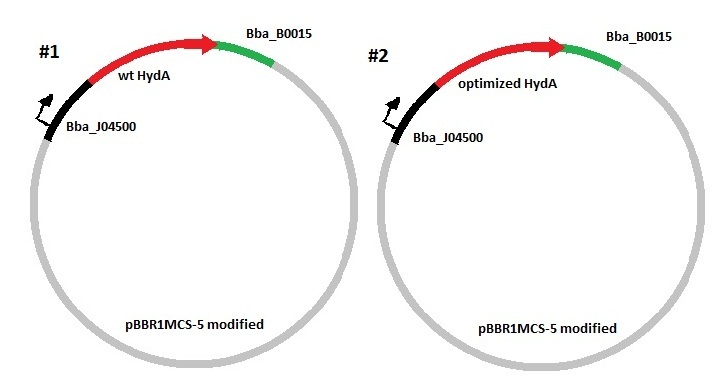
The optimization control is composed of the following parts:
pBBR1MCS-5 derived plasmid. This plasmid is a modification of the pBBR1MCS-5 plasmid, which is a broad-host-range (bhr) vector designed to assist genetic anaylsis in prokaryotes, specifically for Gram- bacteria that are naturally CmR (chloramphenicol resistant). It is used to transfer genes from E. coli to R. etli by first transforming E. coli S17 competent cells with this vector containing an RFP reporter and later performing a conjugation between the transformed E. coli cells and R. etli CFN42.
Bba_J04500. This biopart will permit constitutive expression of the wild-type and the optimized sequences to be tested. This part is composed of a promoter regulated by LacI and a strong RBS. As the host were this part will be found does not contain the LacI repressor, a constitutive expression is therefore expected.
Bba_B0015. This biopart is a double transcriptional terminator consisting of BBa_B0010 and BBa_B0012. It has a high forward efficieny, it will ensure that the growth phenotype that could be observed in the host is due exclusively to the translation of the coding region and not to the excessive transcription from the plasmid pBBR1MCS-5.
HydA wild-type CDS. This sequence belongs to the hydrogenase I gene from Clostridium acetobutylicum ATCC 824. It is 1749 nucleotides long.
HydA optimized CDS. This sequence was made by the bioinformatic tools we developed that change all the codons in the sequence by the most used codons by the constitutively expressed genes in Rhizobium etli CFN42. It is 1749 nucleotide long.
Two constructions are built, each composed of the vector pBBR1MCS-5, the part Bba_J04500, the part Bba_B0015 and differing in the coding region each one possesses, either the wild-type or the optimized hydrogenase I, HydA gene.
Measurement
Once assembled, the plasmids should be transformed to S17 E. coli cells, and latter conjugated to Rhizobium etli CFN42. Once in its final host, both R. etli will be put to test their growing performances. As the one carrying the plasmids that has the optimized version of the hydrogenase, HydA, has the appropiate codons that allow the cell to overexpress the gene without effectively affecting the pool of tRNAs existing within the bacteria, it is expected to be the one with the better growth curves; compared to the one that carries the wild-type version of HydA. We've chose just to test the optimization control with just one gene, and specially with the hydrogenase gene, as we are not expecting to produce molecular hydrogen and because we do not want to affect the growth of the bacteria by using R. etli metabolites with the system. The ferredoxin and the PFOR were not added to this system. Even so, we cannot completely asure that there will not be hydrogen production, as other endogenous redox potential donnors might interact with the hydrogenase.
Justification
Our main objective with the project Hydrobium etli is the production of molecular hydrogen that is profitable to use it as an energy carrier. In order to achieve this, we must ensure that the system we've introduced into Rhizobium etli functions at its best conditions, firing on all cyllinders; but at the same time without affecting the bacterium host. This last implication is of utmost importance, as the bacteria R. etli is symbiont of the plant Phaseolus vulgaris. The health of the bacteria is necessary for the very plant survival.
References
- Paul M.Sharpl and Wen-Hsiung Li. The codon adaptation index - a measure of directional synonymous codon usage bias, and its potential application. Nucleic Acids Research. 1987.
- Grzegorz Kudla, Andrew W. Murray, David Tollervey, Joshua B. Plotkin. Coding-Sequence Determinants of Gene Expression in Escherichia coli. Sicence. 2009.
- Mark Welch Sridhar Govindarajan, Jon E. Ness, Alan Villalobos, Austin Gurney, Jeremy Minshull, Claes Gustafsson. Design Parameters to Control Synthetic Gene Expression in Escherichia coli. PLoS one, 2009
- Sivan Navon, Yitzhak Pilpel. The role of codon selection in regulation of translation efficiency deduced from synthetic libraries. Genome Biology. 2011
- Kuchenreuther JM, Grady-Smith CS, Bingham AS, George SJ, Cramer SP, et al. (2010) High-Yield Expression of Heterologous [FeFe] Hydrogenases in Escherichia coli. PLoS ONE 5(11): e15491
- Agapakis et al.: Insulation of a synthetic hydrogen metabolism circuit in bacteria. Journal of Biological Engineering 2010 4:3.
 "
"