Team:UNAM-Genomics Mexico/Project/HydrogenProduction
From 2011.igem.org
| (10 intermediate revisions not shown) | |||
| Line 11: | Line 11: | ||
__TOC__ | __TOC__ | ||
| + | ==Abstract== | ||
| - | + | Hydrogen production by FeFe hydrogenases requires an anaerobic niche. Taking advantage of microaerobic niche of beans' nodules (little spheres in the roots of beans) and a natural chassis living there (Rhizobium etli), we attempted to couple Hydrogen production into Rhizobium. | |
| + | We kept in mind during the project to take care about nitrogen fixation (natural process by Rhizobium etli in nodules) [[Team:UNAM-Genomics_Mexico/Modeling/FBA| avoiding negative crosstalk]] as well as of avoiding uptake of molecular resources (CAI and DG) needed by Rhizobium etili during symbiosis with beans. | ||
| - | |||
| - | == | + | ==Description== |
| - | + | For the design of our parts we took inspiration from the work done in the Silver Lab in HMS, and expanded its use beyond traditional ''E. coli'' centric views. The system is divided in two operons: | |
| - | |||
| - | + | ===Operon 1=== | |
| + | It Integrates the following elements: | ||
| - | + | *'''Translational fusion of a hydrogenase with a ferrodoxin:''' These two genes (HydA and FeOx) comes from the bacteria ''Clostridium acetobutylicum'' ATCC 824. The hydrogenase HydA1 requires electrons to reduce protons and form hydrogen gas molecules. The ferredoxin is the protein that mediates the electron transfer between an electron transfer donor and a hydrogenase. The ferredoxin and the hydrogenase need to physically interact for the circuit to function; Until now, both proteins' interaction surface has been extensively modeled in silico, with evidence for a strong electrostatic component in this interaction. The translational fusion enhances interaction efficiency between the proteins. | |
| - | + | *'''Pyruvate-ferredoxin oxidoreductase:''' This gene (PFOR) comes from ''Desulfovibrio africanus''. Pyruvate-ferredoxin oxidoreductases, generally catalyze the crucial energy-yielding reaction of pyruvate oxidation and serve as electron carriers. Sometimes they differ among distinct species in their acceptor/donor specificity, substrate specificity, sensitivity to O2, and kinetics of CO2 production and consumption. In this system PFOR acts reducing ferrodoxin as it breaks down pyruvate to obtain Acetyl-CoA. | |
| - | [[File:Unamgenomicsoperon1.jpg|center|Operon1]] | + | In our project we intend to reproduce a pathway with heterologously expressed PFOR-ferredoxin-hydrogenase in order to couple the hydrogen production to the glycolysis process. |
| + | |||
| + | |||
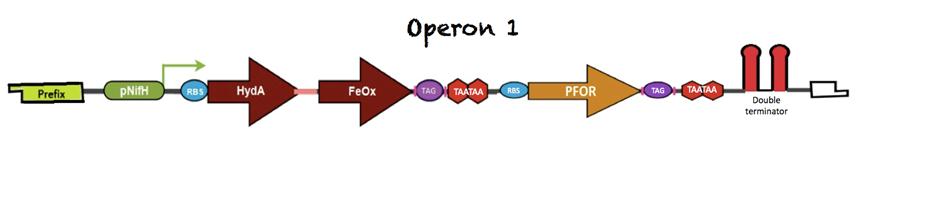
| + | These three sequences are arranged in one operon expressed under a NifH* promoter, which is active under the nitrogen fixation conditions that include low oxygen concentration. The figure below shows the arrangement of the first operon: | ||
| + | |||
| + | |||
| + | |||
| + | [[File:Unamgenomicsoperon1.jpg|850px|center|Operon1]] | ||
| + | |||
This operon starts with a Prefix region, followed by a linker and the pNifH promoter. Downstream of the transcriptional start site there is a linker and a ribosomal binding site (RBS). The fusion between HydA1 and FeOx is mediated by a 14aa long glycine/serine rich linker. A six amino acid long polyhistidine tag has been added at the C-terminus, before the stop codons; this tag is flanked by two HindIII restriction sites. | This operon starts with a Prefix region, followed by a linker and the pNifH promoter. Downstream of the transcriptional start site there is a linker and a ribosomal binding site (RBS). The fusion between HydA1 and FeOx is mediated by a 14aa long glycine/serine rich linker. A six amino acid long polyhistidine tag has been added at the C-terminus, before the stop codons; this tag is flanked by two HindIII restriction sites. | ||
| + | |||
Between the fusion and PFOR there is a spacer and another RBS that precedes PFOR region. A six amino acid long polyhistidine tag (to test presence) and two TAA stop codons had been added at the end of the coding region followed by a double terminator that delimits the end of this first operon. | Between the fusion and PFOR there is a spacer and another RBS that precedes PFOR region. A six amino acid long polyhistidine tag (to test presence) and two TAA stop codons had been added at the end of the coding region followed by a double terminator that delimits the end of this first operon. | ||
| - | *'''NifH promoter:''' We decided to use this promoter region in our project to promote the transcription of gene constructions that, according to our design, need to be expressed in low oxygen conditions in R. etli. The genes regulated by this promoter region will generally be active in the bacteroid state of R. etli because there are low concentrations of oxygen in the nodule’s environment. | + | *'''NifH promoter:''' We decided to use this promoter region in our project to promote the transcription of gene constructions that, according to our design, need to be expressed in low oxygen conditions in ''R. etli''. The genes regulated by this promoter region will generally be active in the bacteroid state of ''R. etli'' because there are low concentrations of oxygen in the nodule’s environment. |
| - | Operon 2 | + | ===Operon 2=== |
| + | |||
Because of the complexity of hydrogenases’ active sites, they require maturation machineries. It is only very recently that the maturation of the active site of their [FeFe] counterparts has been partially elucidated. Three gene products, called HydE, HydF and HydG were found to be necessary and sufficient to mature the HydA hydrogenase. This products function in the assembly of [Fe] hydrogenase, they are accessory genes required for the maturation of an active [Fe] hydrogenase, that’s why they are called "maturases". | Because of the complexity of hydrogenases’ active sites, they require maturation machineries. It is only very recently that the maturation of the active site of their [FeFe] counterparts has been partially elucidated. Three gene products, called HydE, HydF and HydG were found to be necessary and sufficient to mature the HydA hydrogenase. This products function in the assembly of [Fe] hydrogenase, they are accessory genes required for the maturation of an active [Fe] hydrogenase, that’s why they are called "maturases". | ||
| Line 51: | Line 62: | ||
| - | *'''HydEF:''' Is a protein from Chlamydomonas reinhardtii genome, this protein contains two unique domains that are homologous to two distinct prokaryotic proteins, HydE and HydF, which are found exclusively in organisms containing [Fe] hydrogenase. | + | *'''HydEF:''' Is a protein from ''Chlamydomonas reinhardtii'' genome, this protein contains two unique domains that are homologous to two distinct prokaryotic proteins, HydE and HydF, which are found exclusively in organisms containing [Fe] hydrogenase. |
| - | *'''HydG:''' Also comes from Chlamydomonas reinhardtii genome. Specifically HydG is a bi-functional enzyme that uses a relatively simple substrate to generate the triple-bonded ligands of [FeFe]-hydrogenase active site. | + | *'''HydG:''' Also comes from the ''Chlamydomonas reinhardtii'' genome. Specifically HydG is a bi-functional enzyme that uses a relatively simple substrate to generate the triple-bonded ligands of [FeFe]-hydrogenase active site. |
| + | |||
All organisms with [Fe] hydrogenase and sequenced genomes contain homologues of HydE, HydF, and HydG. They belong to the Radical S-adenosylmethionine (designated "Radical SAM", also known as the AdoMet radical) superfamily. There is precedent for the involvement of a Radical SAM protein in the donation of iron to the catalytic metal cluster of an [Fe]-metalloenzyme, thats why its proposed that HydEF and/or HydG proteins play a similar role in the mobilization of iron for assembly of the [Fe] hydrogenase H-cluster. | All organisms with [Fe] hydrogenase and sequenced genomes contain homologues of HydE, HydF, and HydG. They belong to the Radical S-adenosylmethionine (designated "Radical SAM", also known as the AdoMet radical) superfamily. There is precedent for the involvement of a Radical SAM protein in the donation of iron to the catalytic metal cluster of an [Fe]-metalloenzyme, thats why its proposed that HydEF and/or HydG proteins play a similar role in the mobilization of iron for assembly of the [Fe] hydrogenase H-cluster. | ||
| Line 60: | Line 72: | ||
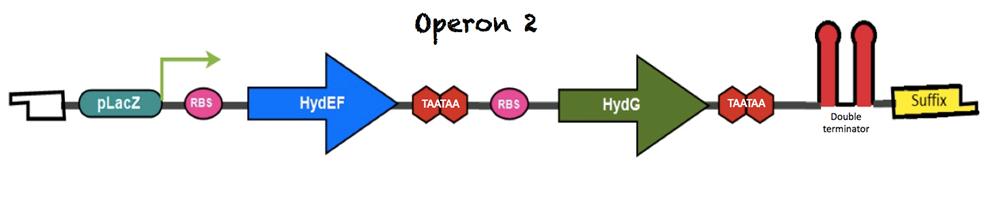
The figure below shows the arrangement of the second operon: | The figure below shows the arrangement of the second operon: | ||
| - | [[File:Unamgenomicsoperon2.jpg|center|Operon2]] | + | |
| + | |||
| + | [[File:Unamgenomicsoperon2.jpg|850px|center|Operon2]] | ||
| + | |||
| + | |||
| + | |||
| + | Operon 2 is expressed under a constitutive promoter LacZ*, downstream of the transcriptional start site there is a linker, a ribosomal binding site (RBS) and another linker that precedes HydEF gene. Two TAA stop codons had been added at the end of the coding region. | ||
| + | |||
| + | |||
| + | Between HydEF and HydG there is a spacer and another RBS for HydG translation. Two TAA stop codons had been added at the end of the coding region followed by a double terminator that delimits the end of the second operon; a Suffix region is next to this terminator. | ||
| + | |||
| + | |||
| + | *'''LacZ promoter:''' We used this promoter in order to have a considerable previous concentration of maturases before hypoxic conditions are induced for expression of hydrogenase. This assures a rapidly production of hydrogenase in response to the low oxygen conditions. | ||
| + | |||
| + | |||
| + | These two operons are going to be integrated in the same plasmid, one next to the other conforming one big fragment, flanked by prefix and suffix regions, comprising about 11.8 kb. First is going to be transformed in an ''E. coli'' strain so we could later conjugate it with ''Rhizobium etli'' and test the system operation. | ||
| + | |||
| + | |||
| + | |||
| + | Consult our logbook for the assembly of this system [[Team:UNAM-Genomics_Mexico/Notebook/SA|here]]. | ||
| + | |||
| + | ==Justification== | ||
| + | |||
| + | Whit this construction and [[Team:UNAM-Genomics_Mexico/Project/RhizobialKit|Rhizobial Kit]] we purpose a R. etli as a good self-contained system. This construction is coupled with Rhizobium physiology at Symbiosis moment in the nodules, yielding Hidrogen production and keeping Nitrogen Fixation. Many parts of this system have been used by other groups, and we emphisize the potencial of natural chassis and self-contained systems for synthetic biology. | ||
| + | |||
| + | |||
| + | |||
| + | ==Reference:== | ||
| + | |||
| + | Agapakis et al.: Insulation of a synthetic hydrogen metabolism circuit in bacteria. Journal of Biological Engineering 2010 4:3. | ||
| + | |||
| + | Kuchenreuther JM, Grady-Smith CS, Bingham AS, George SJ, Cramer SP, et al. (2010) High-Yield Expression of Heterologous [FeFe] Hydrogenases in ''Escherichia coli''. PLoS ONE 5(11): e15491 | ||
| + | |||
| + | Nicolet Y, Martin L, Tron C, Fontecilla-Camps JC (2010) A glycyl free radical as the precursor in the synthesis of carbon monoxide and cyanide by the [FeFe]-hydrogenase maturase HydG. FEBS Lett 584: 4197–4202. | ||
| + | |||
| + | |||
| + | Pieulle, L., Guigliarelli, B., Asso, M., Dole, F., Bernadac, A., and Hatchikian, E. C. (1995) Isolation and characterization of the pyruvate:ferredoxin oxidoreductase from the sulfate-reducing bacterium ''Desulfovibrio africanus'', Biochim. Biophys. Acta 1250, 49-59. | ||
| + | |||
| + | Posewitz, M. C., King, P. W., Smolinski, S. L., Zhang, L., Seibert, M. & Ghirardi, M. L. (2004). Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active [Fe] hydrogenase. J Biol Chem 279, 25711–25720. | ||
}} | }} | ||
Latest revision as of 02:57, 29 September 2011
Hydrogen Production
Contents |
Abstract
Hydrogen production by FeFe hydrogenases requires an anaerobic niche. Taking advantage of microaerobic niche of beans' nodules (little spheres in the roots of beans) and a natural chassis living there (Rhizobium etli), we attempted to couple Hydrogen production into Rhizobium. We kept in mind during the project to take care about nitrogen fixation (natural process by Rhizobium etli in nodules) avoiding negative crosstalk as well as of avoiding uptake of molecular resources (CAI and DG) needed by Rhizobium etili during symbiosis with beans.
Description
For the design of our parts we took inspiration from the work done in the Silver Lab in HMS, and expanded its use beyond traditional E. coli centric views. The system is divided in two operons:
Operon 1
It Integrates the following elements:
- Translational fusion of a hydrogenase with a ferrodoxin: These two genes (HydA and FeOx) comes from the bacteria Clostridium acetobutylicum ATCC 824. The hydrogenase HydA1 requires electrons to reduce protons and form hydrogen gas molecules. The ferredoxin is the protein that mediates the electron transfer between an electron transfer donor and a hydrogenase. The ferredoxin and the hydrogenase need to physically interact for the circuit to function; Until now, both proteins' interaction surface has been extensively modeled in silico, with evidence for a strong electrostatic component in this interaction. The translational fusion enhances interaction efficiency between the proteins.
- Pyruvate-ferredoxin oxidoreductase: This gene (PFOR) comes from Desulfovibrio africanus. Pyruvate-ferredoxin oxidoreductases, generally catalyze the crucial energy-yielding reaction of pyruvate oxidation and serve as electron carriers. Sometimes they differ among distinct species in their acceptor/donor specificity, substrate specificity, sensitivity to O2, and kinetics of CO2 production and consumption. In this system PFOR acts reducing ferrodoxin as it breaks down pyruvate to obtain Acetyl-CoA.
In our project we intend to reproduce a pathway with heterologously expressed PFOR-ferredoxin-hydrogenase in order to couple the hydrogen production to the glycolysis process.
These three sequences are arranged in one operon expressed under a NifH* promoter, which is active under the nitrogen fixation conditions that include low oxygen concentration. The figure below shows the arrangement of the first operon:
This operon starts with a Prefix region, followed by a linker and the pNifH promoter. Downstream of the transcriptional start site there is a linker and a ribosomal binding site (RBS). The fusion between HydA1 and FeOx is mediated by a 14aa long glycine/serine rich linker. A six amino acid long polyhistidine tag has been added at the C-terminus, before the stop codons; this tag is flanked by two HindIII restriction sites.
Between the fusion and PFOR there is a spacer and another RBS that precedes PFOR region. A six amino acid long polyhistidine tag (to test presence) and two TAA stop codons had been added at the end of the coding region followed by a double terminator that delimits the end of this first operon.
- NifH promoter: We decided to use this promoter region in our project to promote the transcription of gene constructions that, according to our design, need to be expressed in low oxygen conditions in R. etli. The genes regulated by this promoter region will generally be active in the bacteroid state of R. etli because there are low concentrations of oxygen in the nodule’s environment.
Operon 2
Because of the complexity of hydrogenases’ active sites, they require maturation machineries. It is only very recently that the maturation of the active site of their [FeFe] counterparts has been partially elucidated. Three gene products, called HydE, HydF and HydG were found to be necessary and sufficient to mature the HydA hydrogenase. This products function in the assembly of [Fe] hydrogenase, they are accessory genes required for the maturation of an active [Fe] hydrogenase, that’s why they are called "maturases".
According to this, the second operon is composed of the following elements:
- HydEF: Is a protein from Chlamydomonas reinhardtii genome, this protein contains two unique domains that are homologous to two distinct prokaryotic proteins, HydE and HydF, which are found exclusively in organisms containing [Fe] hydrogenase.
- HydG: Also comes from the Chlamydomonas reinhardtii genome. Specifically HydG is a bi-functional enzyme that uses a relatively simple substrate to generate the triple-bonded ligands of [FeFe]-hydrogenase active site.
All organisms with [Fe] hydrogenase and sequenced genomes contain homologues of HydE, HydF, and HydG. They belong to the Radical S-adenosylmethionine (designated "Radical SAM", also known as the AdoMet radical) superfamily. There is precedent for the involvement of a Radical SAM protein in the donation of iron to the catalytic metal cluster of an [Fe]-metalloenzyme, thats why its proposed that HydEF and/or HydG proteins play a similar role in the mobilization of iron for assembly of the [Fe] hydrogenase H-cluster.
The figure below shows the arrangement of the second operon:
Operon 2 is expressed under a constitutive promoter LacZ*, downstream of the transcriptional start site there is a linker, a ribosomal binding site (RBS) and another linker that precedes HydEF gene. Two TAA stop codons had been added at the end of the coding region.
Between HydEF and HydG there is a spacer and another RBS for HydG translation. Two TAA stop codons had been added at the end of the coding region followed by a double terminator that delimits the end of the second operon; a Suffix region is next to this terminator.
- LacZ promoter: We used this promoter in order to have a considerable previous concentration of maturases before hypoxic conditions are induced for expression of hydrogenase. This assures a rapidly production of hydrogenase in response to the low oxygen conditions.
These two operons are going to be integrated in the same plasmid, one next to the other conforming one big fragment, flanked by prefix and suffix regions, comprising about 11.8 kb. First is going to be transformed in an E. coli strain so we could later conjugate it with Rhizobium etli and test the system operation.
Consult our logbook for the assembly of this system here.
Justification
Whit this construction and Rhizobial Kit we purpose a R. etli as a good self-contained system. This construction is coupled with Rhizobium physiology at Symbiosis moment in the nodules, yielding Hidrogen production and keeping Nitrogen Fixation. Many parts of this system have been used by other groups, and we emphisize the potencial of natural chassis and self-contained systems for synthetic biology.
Reference:
Agapakis et al.: Insulation of a synthetic hydrogen metabolism circuit in bacteria. Journal of Biological Engineering 2010 4:3.
Kuchenreuther JM, Grady-Smith CS, Bingham AS, George SJ, Cramer SP, et al. (2010) High-Yield Expression of Heterologous [FeFe] Hydrogenases in Escherichia coli. PLoS ONE 5(11): e15491
Nicolet Y, Martin L, Tron C, Fontecilla-Camps JC (2010) A glycyl free radical as the precursor in the synthesis of carbon monoxide and cyanide by the [FeFe]-hydrogenase maturase HydG. FEBS Lett 584: 4197–4202.
Pieulle, L., Guigliarelli, B., Asso, M., Dole, F., Bernadac, A., and Hatchikian, E. C. (1995) Isolation and characterization of the pyruvate:ferredoxin oxidoreductase from the sulfate-reducing bacterium Desulfovibrio africanus, Biochim. Biophys. Acta 1250, 49-59.
Posewitz, M. C., King, P. W., Smolinski, S. L., Zhang, L., Seibert, M. & Ghirardi, M. L. (2004). Discovery of two novel radical S-adenosylmethionine proteins required for the assembly of an active [Fe] hydrogenase. J Biol Chem 279, 25711–25720.
 "
"