Team:UNAM-Genomics Mexico/Notebook/SA
From 2011.igem.org
| (6 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:UNAM-Genomics Mexico/Templates/Modeling| content= | {{:Team:UNAM-Genomics Mexico/Templates/Modeling| content= | ||
__NOTOC__ | __NOTOC__ | ||
| - | |||
| - | |||
| - | |||
=Lab Logbook - System Assembling.= | =Lab Logbook - System Assembling.= | ||
| Line 13: | Line 10: | ||
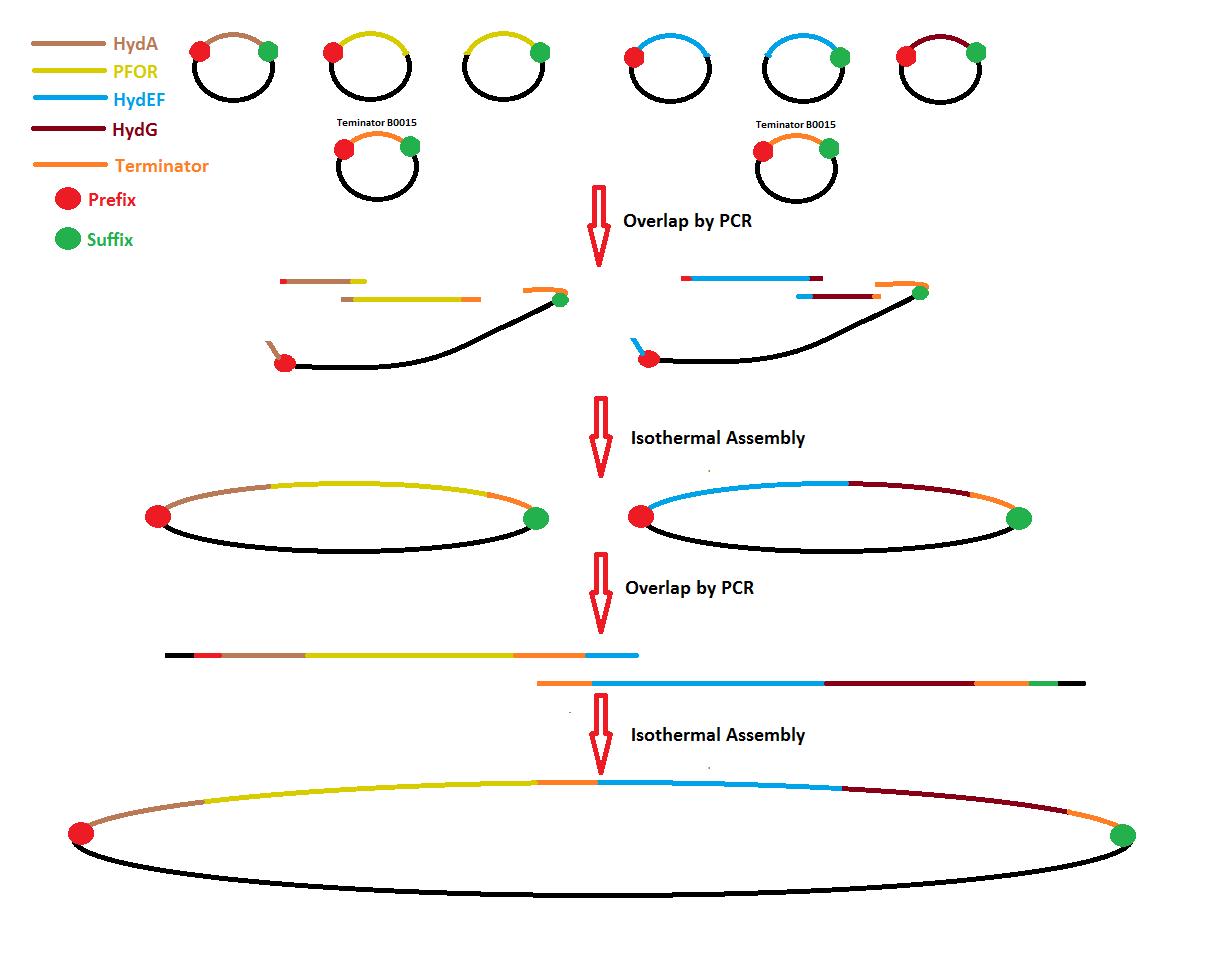
[[File:Unamgenomicssafig1.jpg|900px|center|Fig. 1 General overview of system assembly.]] | [[File:Unamgenomicssafig1.jpg|900px|center|Fig. 1 General overview of system assembly.]] | ||
| + | |||
| + | |||
| + | __TOC__ | ||
| Line 18: | Line 18: | ||
==Week 1== | ==Week 1== | ||
| - | 2nd - 8th September | + | ===2nd - 8th September=== |
| - | + | ||
| + | After 2 months of delay, DNA synthesis from Mr. Gene has arrived at 22:00 hrs. They are 6 syntheses HydA, PFOR1, PFOR2, HydEF1, HydEF2 and HydG. PFORs and HydEFs were synthesized separately because sequences are too long for synthesis. | ||
| - | |||
| - | + | #With this arrival it is necessary to get regions for Isothermal Assembly with PCR to make overlap between sequences. | |
| - | + | #Trasnsform cells with HydA and PFOR1 to get plasmid and split a Periplasm Tag (PT) designed by us. | |
| + | |||
| + | #PCR for Terminator-backbone (Ter-Back) in Isothermal Assembly. | ||
| Line 52: | Line 53: | ||
==Week 2== | ==Week 2== | ||
| - | 9th -15th September | + | ==9th -15th September=== |
| + | |||
Repeating successful PCRs for Ter-Back1 and try to get Ter-Back2 but both failed. | Repeating successful PCRs for Ter-Back1 and try to get Ter-Back2 but both failed. | ||
| Line 62: | Line 64: | ||
1-Ladder 500bp 2-HydEF2 55°C 3-HydEF2 60°C 4-HydEF2 62°C 5-HydEF1 55°C 6-HydEF1 60°C 7-HydEF1 62°C 8-HydG 55°C 9-PFOR2 55°C 10-HydG 60°C 11-HydG 62°C 12-PFOR2 60°C 13-PFOR2 62°C | 1-Ladder 500bp 2-HydEF2 55°C 3-HydEF2 60°C 4-HydEF2 62°C 5-HydEF1 55°C 6-HydEF1 60°C 7-HydEF1 62°C 8-HydG 55°C 9-PFOR2 55°C 10-HydG 60°C 11-HydG 62°C 12-PFOR2 60°C 13-PFOR2 62°C | ||
| - | Band | + | Band isolation for HydG and PFRO2 was performed, but there are remaining of DNA template. |
| Line 68: | Line 70: | ||
==Week 3== | ==Week 3== | ||
| - | 16th-24th September | + | ===16th-24th September=== |
A second band extraction was performed to get bona fide PCR products. | A second band extraction was performed to get bona fide PCR products. | ||
| Line 79: | Line 81: | ||
| + | See the description of this system [[Team:UNAM-Genomics_Mexico/Project/HydrogenProduction|here]]. | ||
}} | }} | ||
Latest revision as of 23:13, 28 September 2011
Lab Logbook - System Assembling.
Process overview
Contents |
Week 1
2nd - 8th September
After 2 months of delay, DNA synthesis from Mr. Gene has arrived at 22:00 hrs. They are 6 syntheses HydA, PFOR1, PFOR2, HydEF1, HydEF2 and HydG. PFORs and HydEFs were synthesized separately because sequences are too long for synthesis.
- With this arrival it is necessary to get regions for Isothermal Assembly with PCR to make overlap between sequences.
- Trasnsform cells with HydA and PFOR1 to get plasmid and split a Periplasm Tag (PT) designed by us.
- PCR for Terminator-backbone (Ter-Back) in Isothermal Assembly.
First attempt failed but not at all, just PFOR_Dos and hydG amplified and positive control, for that we need a troubleshooting for PCR. The same case was for Ter-Back . There is a slight amplification of HydEF1.
First attempt in troubleshooting for HydEFs failed, this time we use other Polymerase (Taq Platinum) and didn´t work. Changing annealing temperatures we had a possible way to get HydEF2.
Plasmid extraction of HydA and PFOR was made.
Digestion with NdeI to Split PT was done, but results are confusing, because self Ligation didn´t work.
In order to do a band extraction we need more PCR product to successful PCRs we amplify them, band extraction was polluted by template DNA.
There is a possible way to get HydEF2, is necessary check that and perform more PCRs for more product. Ter-Back 1 amplified but Ter-Back2 didn´t.
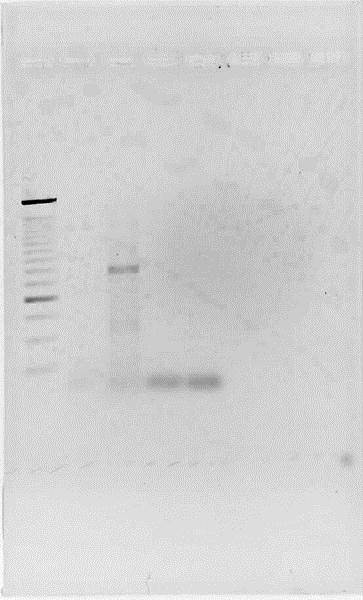
Fig.2. Successful PCR of Terminator 1.
Lanes 1. Ladder 500bp 2. Ter-Back 1 3. Ter-Back 2 4.HydEF 1 5. HydEF 2
Week 2
9th -15th September=
Repeating successful PCRs for Ter-Back1 and try to get Ter-Back2 but both failed.
Giving more time in Initial Denaturalization (5:00 min) we got better results, even for difficult PCRs as HydEF1 and different annealing temperatures
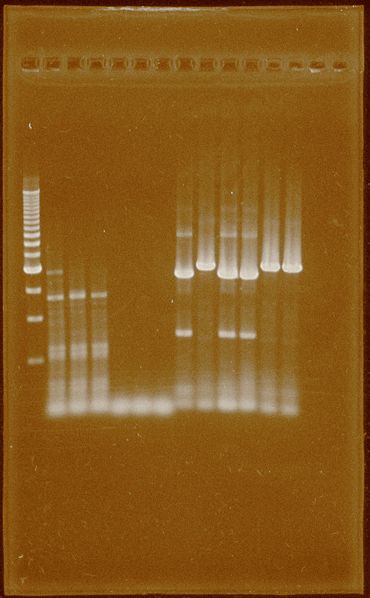
Fig.3 Successful PCR for PFOR2 and HYDEF2, each three lanes. 1-Ladder 500bp 2-HydEF2 55°C 3-HydEF2 60°C 4-HydEF2 62°C 5-HydEF1 55°C 6-HydEF1 60°C 7-HydEF1 62°C 8-HydG 55°C 9-PFOR2 55°C 10-HydG 60°C 11-HydG 62°C 12-PFOR2 60°C 13-PFOR2 62°C
Band isolation for HydG and PFRO2 was performed, but there are remaining of DNA template.
Week 3
16th-24th September
A second band extraction was performed to get bona fide PCR products.
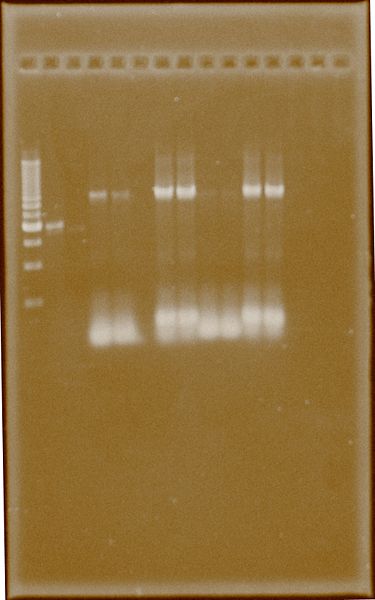
Ter-Back 2 was amplified to get more PCR product, and with the same instructions Ter-Back 1 appears slightly amplified. The difference lies in addition of DMSO, probably the given nature of Terminator need it.
Fig.4. Numbers (1) and (2) represents 54.7°C and 60.3 °C for annealing respectively. 1. Ladder 500bp 2. PFOR2 E.B.‡ 3. HydG E.B.‡ 4. Ter-Back1 DMSO (1) 5. Ter-Back1 DMSO (2) 6. Empty 7. Ter-Back2 DMSO (1) 8. Ter-Back2 DMSO (2) 9. Ter-Back1 Not DMSO (1) 10. Ter-Back1 Not DMSO (2) 11. Ter-Back2 Not DMSO (1) 12. Ter-Back2 Not DMSO (2)
See the description of this system here.
 "
"