Team:Calgary/Project/ProjectPseudomonas
From 2011.igem.org
Rpgguardian (Talk | contribs) |
Rpgguardian (Talk | contribs) |
||
| (35 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
{{Team:Calgary/ProjectBar| | {{Team:Calgary/ProjectBar| | ||
| - | TITLE= | + | TITLE=Novel Application of Biotinylation| |
BODY=<html> | BODY=<html> | ||
<img style="float: right; width: 150px; padding-left: 10px; padding-bottom: 5px; padding-top: 5px;" src="https://static.igem.org/mediawiki/2011/0/01/UCalgary2011_PromoterFredFishing.png"></img> | <img style="float: right; width: 150px; padding-left: 10px; padding-bottom: 5px; padding-top: 5px;" src="https://static.igem.org/mediawiki/2011/0/01/UCalgary2011_PromoterFredFishing.png"></img> | ||
| - | <p>Our team set out to identify a novel responsive element capable of detecting and quantifying naphthenic acids ( | + | <p>Our team set out to identify a novel responsive element capable of detecting and quantifying naphthenic acids (NAs) in solution. While numerous studies have begun to identify species of bacteria which can survive and/or degrade NAs, the pathways of degradation have not yet been characterized. Therefore this goal required our team to design and implement novel methods to isolate the cellular systems of tailing ponds biological organisms which detect and potentially degrade naphthenic acids. We developed a naphthenic acid biotinylation/immunoprecipitation technique which allows for identification of novel protein complexes which interact and may potentially detect naphthenic acids. Our team developed an optimized protocol for the biotinylation of cyclohexanepentanoic acid (CHPA), cyclohexanebutyric acid (CHBA), and cyclohexanecarboxylic acid (CHCA), and a protocol for the immunoprecipitation of <i>Pseudomonas</i> lysates using commercially available streptavidin magnetic beads.</p> |
| - | <p>In addition, our team developed an <i>in silico</i> approach to isolating proteins which may be involved in degrading naphthenic acids using two specific species of <i>Pseudomonas</i> previously reported to degrade | + | <p>In addition, our team developed an <i>in silico</i> approach to isolating proteins which may be involved in degrading naphthenic acids using two specific species of <i>Pseudomonas</i> previously reported to degrade NAs in co-culture. Through use of various bioinformatic databases, a list of potential naphthenic acid degrading proteins was identified. Of these, a set were tested using reverse transcriptase polymerase chain reaction (RT-PCR) to identify a change in expression when incubated with naphthenic acids. A novel Enoyl-Coa-hydratase was identified and may be a responsive element to naphthenic acids.</p> |
<p><b><a href="https://2011.igem.org/Team:Calgary/Project/Promoter/Bioinformatics">Click here</a> for more information about our Bioinformatic search.</b></p> | <p><b><a href="https://2011.igem.org/Team:Calgary/Project/Promoter/Bioinformatics">Click here</a> for more information about our Bioinformatic search.</b></p> | ||
| - | <h2> | + | <h2>Sensory Element Fishing: Designing A Novel Biotinylation-Immunprecipitation System For Isolation of Naphthenic Acid Interactors</h2> |
<h2>What Are We Looking For?</h2> | <h2>What Are We Looking For?</h2> | ||
| Line 17: | Line 17: | ||
</html>[[Image:UofC2011_Looking.png|thumb|400px|right|<b>Figure 1</b> The model naphthenic acid Cyclohexanepentanoic acid and the potential targets which our team was looking for in our screen for a NA responsive element. Our biotinylation/immunoprecipitation technique can be optimized for searching for any of these four response elements. The techniques described further in this document focus on transcription factor and degradation enzyme based optimization.]]<html> | </html>[[Image:UofC2011_Looking.png|thumb|400px|right|<b>Figure 1</b> The model naphthenic acid Cyclohexanepentanoic acid and the potential targets which our team was looking for in our screen for a NA responsive element. Our biotinylation/immunoprecipitation technique can be optimized for searching for any of these four response elements. The techniques described further in this document focus on transcription factor and degradation enzyme based optimization.]]<html> | ||
| - | <p>Due to the complexity of biological systems, our team focused our efforts on identifying a system for identification of proteins or complexes which bind DNA. Transcription factors, which bind to specific elements of a gene known as the promoter, have been well characterized in other metabolic systems to respond to metabolites in the cell and change the level of transcription of particular metabolic response genes. A common example of one of these systems is the | + | <p>Due to the complexity of biological systems, our team focused our efforts on identifying a system for identification of proteins or complexes which bind DNA. Transcription factors, which bind to specific elements of a gene known as the promoter, have been well characterized in other metabolic systems to respond to metabolites in the cell and change the level of transcription of particular metabolic response genes. A common example of one of these systems is the Lac operon which has been harnessed as a selection system already in the registry. Identifying a system such as this for naphthenic acids, if one exists, would allow for a specific response to be generated to many different kinds of naphthenic acids. Our hypothesis requires that the organisms we use respond specifically to naphthenic acids and result in specific upregulation of metabolic genes with little background effect in the cell. Additionally, numerous protein elements have been shown to bind and modify DNA, resulting in downstream changes to protein expression. Examples of this are histone modifying enzymes in Eukaryotes which have opened up a new realm of research known as epigenetics. These systems can be complicated and often do not directly recognize metabolic chemical compounds. Furthermore, it is possible that naphthenic acids may even interact with specific RNA responsive elements known as riboswitches, which may be involved in the modification of protein expression. While the identification of any of these systems were of specific interest to us to identify a detection system for naphthenic acids, we wanted our approach to be broad enough to idenitfy any components interacting with naphthenic acids.</p> |
| - | <h2> | + | <h2>Naphthenic Acid Degrading Organisms To Be Used</h2> |
<p>Due to the diversity and ambiguity of microbial species in the tailings ponds, we chose to focus our efforts for our promoter search on two species of <i>Pseudomonas</i> and a species of microalgae <i>Dunaliella tertiolecta</i> (<b>please see our <a href="https://2011.igem.org/Team:Calgary/Project/Chassis">chassis section</a> for more information on these species</b>). All of these organisms have been shown to degrade naphthenic acids, however little work has been done in characterizing the degradation pathways of both species.</p> | <p>Due to the diversity and ambiguity of microbial species in the tailings ponds, we chose to focus our efforts for our promoter search on two species of <i>Pseudomonas</i> and a species of microalgae <i>Dunaliella tertiolecta</i> (<b>please see our <a href="https://2011.igem.org/Team:Calgary/Project/Chassis">chassis section</a> for more information on these species</b>). All of these organisms have been shown to degrade naphthenic acids, however little work has been done in characterizing the degradation pathways of both species.</p> | ||
| Line 28: | Line 28: | ||
</html>[[Image:UofC2011_IP.png|thumb|400px|right|<b>Figure 2</b> A typical immunoprecipitation (IP) performed using an antibody raised against a particular protein target. The antibody is incubated with a cell lysate containing purified protein. The antibody will bind to its specific antigen and the solution may be incubated with beads that are capable of binding the antibodies. The beads, coupled to the antibody and protein target can then be isolated from the mixture through the use of a magnet (if the beads are magnetic) or through centrifugation. The protein can be denatured off of the beads and analyzed using mass spectrometry to identify their sequence.]]<html> | </html>[[Image:UofC2011_IP.png|thumb|400px|right|<b>Figure 2</b> A typical immunoprecipitation (IP) performed using an antibody raised against a particular protein target. The antibody is incubated with a cell lysate containing purified protein. The antibody will bind to its specific antigen and the solution may be incubated with beads that are capable of binding the antibodies. The beads, coupled to the antibody and protein target can then be isolated from the mixture through the use of a magnet (if the beads are magnetic) or through centrifugation. The protein can be denatured off of the beads and analyzed using mass spectrometry to identify their sequence.]]<html> | ||
| - | <p>As we plan to screen both bacterial and microalgae species, it was vital that our screening technique be robust and efficient in a variety of cellular systems. It was also important that it selectively targeted protein elements which were specific for naphthenic acids and did not result in unwanted background. A common technique used in similar | + | <p>As we plan to screen both bacterial and microalgae species, it was vital that our screening technique be robust and efficient in a variety of cellular systems. It was also important that it selectively targeted protein elements which were specific for naphthenic acids and did not result in unwanted background. A common technique used in similar applications including protein biochemistry is immunoprecipitation (IP) for the identification of protein interactors against a particular bait protein. An antibody is incubated with the lysate of the organism of interest. The target of this antibody and its interacting proteins can be isolated using beads that bind to a conserved region opposite the antigen binding domain. Isolation of the target protein along with its interactors allows for their identification through mass spectrometry (MS), a routinely used technique to identify proteins. This technique has been well established and is used routinely in a variety of applications.</p> |
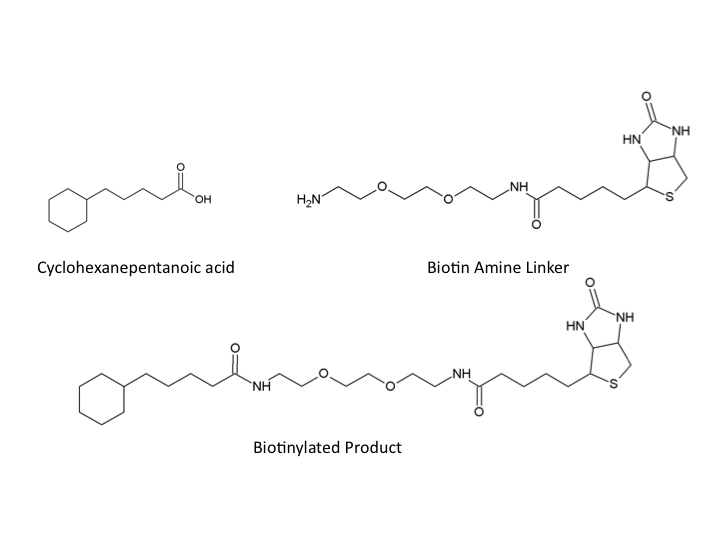
| - | <p>In a similar manner we designed a protocol to use naphthenic acids in an immunoprecipitation. This however, required that we develop a system to conjugate our naphthenic acids to a bead platform similar to the commercially available antibodies used in traditional IP. To accomplish this, we attempted to react the conserved carboxylic acid of the naphthenic acid cyclohexanepentanoic acid (CHPA) (<b>see <a href="https://2011.igem.org/Team:Calgary/Project"> overview section</a></b>) with a commercially available biotin-amine derivative. By reacting these compounds in the presence of EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide) the carboxylic acid and primary amine can produce an amide linkage under low pH conditions. Biotinylation is commonly performed with DNA and protein side chains using commercial kits designed for these applications. However, the biotinylation of small molecules is rare and techniques involving hydrophobic compounds such as CHPA are not found in the literature. This required that we generate a novel protocol for biotinylating CHPA along with other naphthenic acid compounds.</p> | + | <p>In a similar manner we designed a protocol to use naphthenic acids in an immunoprecipitation. This however, required that we develop a system to conjugate our naphthenic acids to a bead platform similar to the commercially available antibodies used in traditional IP. To accomplish this, we attempted to react the conserved carboxylic acid of the naphthenic acid cyclohexanepentanoic acid (CHPA) (<b>see <a href="https://2011.igem.org/Team:Calgary/Project"> overview section</a></b>) with a commercially available biotin-amine derivative. By reacting these compounds in the presence of EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide) the carboxylic acid and primary amine can produce an amide linkage under low pH conditions. Biotinylation is commonly performed with DNA and protein side chains using commercial kits designed for these applications. However, the biotinylation of small molecules is rare and techniques involving hydrophobic compounds such as CHPA are not found in the literature. This required that we generate a <b>novel</b> protocol for biotinylating CHPA along with other naphthenic acid compounds.</p> |
</html>[[Image:UofC2011_Compounds.png|thumb|400px|left|<b>Figure 3</b> Chemical structure of Cyclohexanepentanoic acid (CHPA), Biotin-Amine, and the final product with an amide bonding formed between the carboxylic acid of CHPA and the primary amine group of the Biotin linker.]]<html> | </html>[[Image:UofC2011_Compounds.png|thumb|400px|left|<b>Figure 3</b> Chemical structure of Cyclohexanepentanoic acid (CHPA), Biotin-Amine, and the final product with an amide bonding formed between the carboxylic acid of CHPA and the primary amine group of the Biotin linker.]]<html> | ||
| - | <p>Once our naphthenic acid has become biotinylated, the biotin group will strongly interact with a protein known as streptavidin. The biotin/streptavidin interaction is one of the strongest characterized | + | <p>Once our naphthenic acid has become biotinylated, the biotin group will strongly interact with a protein known as streptavidin. The biotin/streptavidin interaction is one of the strongest binding interactions characterized in biology (Kd=1x10<sup>-15</sup>M) and is commonly cross-linked to magnetic beads allowing for our biotinylated naphthenic acid to be used in similar protocols as in immunoprecipitation. In theory, proteins which interact with the cyclic ring structure of cyclohexane pentanoic acid can be isolated using our biotinylated CHPA/streptavidin beads platform. Finally mass spectrometry can be used to determine the identity of these regulatory elements (See Figure 3).</p> |
<br> | <br> | ||
<h2>Biotinylation</h2> | <h2>Biotinylation</h2> | ||
| + | </html>[[Image:UofC2011_ReactionMech.png|thumb|600px|left|<b>Figure 4</b> Chemical mechanism for the formation of a amide bond between cylohexanepentanoic acid and a commercially available biotin-amine compound.]]<html> | ||
| - | + | <p>The biotinylation of molecules containing carboxylic acid groups follows a well characterized reaction mechanism. In the context of our biotinylation reagents (CHPA and Biotin-PEG-amine) the reaction mechanism can be seen in Figure 4. The cyclohexanepentanoic acid will react with the carbodiimide (EDC) to produce an O-acylisourea intermediate activating the carboxylic acid making it an excellent leaving group. This requires a low pH environment to deprotonate the carboxylic acid. From this intermediate the desired product can be produced via direct attack of the negatively charged carboxylic acid with the amine group of our biotinylated compound producing a urea based compound as a by-product. Further reaction of the O-acylisourea intermediate with cyclohexanepentanoic acid may result in an acid anhydride which then produces the biotinylated compound of interest. The main byproduct of the reaction involves the rearrangement of the O-acylisourea to a stable N-acylurea. Common solutions to reduce the reaction towards this product involves the use of low-dielectric constant solvents such as dichloromethane. Due to the hydrophobic nature of cyclohexanepentanoic acid, aqueous low pH solutions will cause precipitation of this reaction substrate, therefore, the use of other solvents such as dimethylsulfoxide need to be used. The final reaction solution used in all biotinylation experiments used 9% DCM/ 18% ddH2O / 73% DMSO. Typical reactions run overnight at room temperature. (Please see our protocols page for more information on the biotinylation experiment).</p> | |
| - | + | ||
| - | <p>The biotinylation of molecules containing | + | |
<h2>Biotinylation Confirmation</h2> | <h2>Biotinylation Confirmation</h2> | ||
| - | <p>In order to confirm the biotinylation reaction was occurring, reverse phase High Performance Liquid Chromatography (HPLC) was used. A C18 analytical column | + | <p>In order to confirm the biotinylation reaction was occurring, reverse phase High Performance Liquid Chromatography (HPLC) was used. A C18 analytical column (Dionex Acclaim 120) connected to a Dionex ICS-5000 series HPLC was adapted from a program which was based on complex naphthenic acid solution.</p> |
| - | <p>The HPLC conditions used for the analysis | + | <p>The HPLC conditions used for the analysis were as follows. A 60 min. program was designed with a mobile phase of 40% Acetonitrile / 60% 0.1% TFA Water with a gradient from 3-28 min. changing to 85% Acetonitrile/15% 0.1% TFA Water solution holding until 40 min. The final 5-20 min. of the program (runs later in the year were modulated such that there was only a 5 minute ramp back to starting conditions) ramped back to original gradient to clean the column. A 25 μL sample loop was used for all injections except for those used in purifying large amounts of biotinylated compound where a 500 μL sample loop was used. Samples were loaded onto the column at a flow rate of 0.6 mL/min at a temperature of 30<degree>C. |
<p>To establish a set of control conditions for this data analysis, DMSO and DCM were individually injected onto the column and a read out at 210 nm on a UV/VIS spectrophotometer over a 60 min.</p> | <p>To establish a set of control conditions for this data analysis, DMSO and DCM were individually injected onto the column and a read out at 210 nm on a UV/VIS spectrophotometer over a 60 min.</p> | ||
| Line 71: | Line 70: | ||
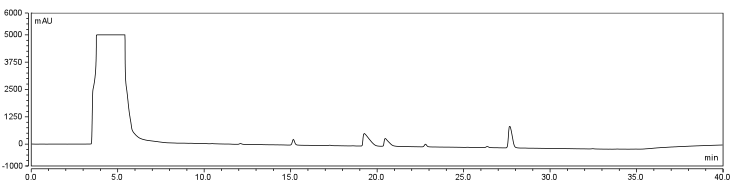
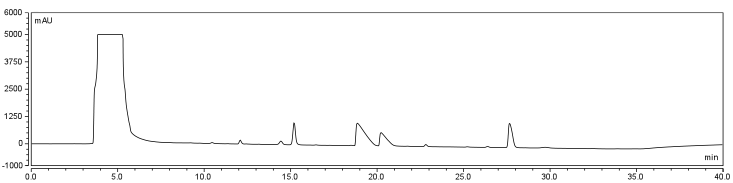
</html>[[Image:UofC2011_DCMRxn.png|thumb|625px|center|<b>Figure 7.b</b> Biotinylation reaction in DCM/DMSO/Water. Samples were incubated at room temperature overnight and injected onto a C18 column using regular protocol.]]<html> | </html>[[Image:UofC2011_DCMRxn.png|thumb|625px|center|<b>Figure 7.b</b> Biotinylation reaction in DCM/DMSO/Water. Samples were incubated at room temperature overnight and injected onto a C18 column using regular protocol.]]<html> | ||
| - | <p>Comparison of the two datasets shows an increase in the three reaction peaks with relatively no change in the CHPA peak, suggesting that dichloromethane’s low dielectric constant increases the relative rate of reaction. Finally, to confirm the identity of our biotinylated product, peak fractions were collected for a potential biotin peak (~ | + | <p>Comparison of the two datasets shows an increase in the three reaction peaks with relatively no change in the CHPA peak, suggesting that dichloromethane’s low dielectric constant increases the relative rate of reaction. Finally, to confirm the identity of our biotinylated product, peak fractions were collected for a potential biotin peak (~4 min.), the peak at 19 min. and the second peak at 20 min. The expected masses of biotin control and the expected products are as follows: |
<ul> | <ul> | ||
| Line 81: | Line 80: | ||
Samples were collected from the HPLC in 30 sec. fractions of 0.3mL and dried down in a vacufuge at 45C. Samples were submitted to the University of Calgary Department of Chemistry MS Facility for electrospray analysis. | Samples were collected from the HPLC in 30 sec. fractions of 0.3mL and dried down in a vacufuge at 45C. Samples were submitted to the University of Calgary Department of Chemistry MS Facility for electrospray analysis. | ||
| - | Figure | + | </p> |
| + | </html>[[Image:UofC2011_MSData2.png|thumb|625px|center|<b>Figure 8</b> Mass Spectrometry Electrospray data for the three peaks with retention times (from left to right) of 4 min. (Control Biotin), 19 min. (The Biotinylated Product), and 20 min. (N-acylurea side reaction). Please click on enlarge to view a better quality version of this image.]]<html> | ||
| - | <p>MS analysis identified the control Biotin at the correct size as determined by the literature. The peak at 19 min. corresponds to the size of Biotin-CHPA plus a sodium ion (22.99 Da) which is found in high quanitites in the methanol used in electrospray which may be responsible for the higher peak size identified. Finally, the peak at 20 min. corresponded to the N-acylurea, the by-product of the reaction. This was an interesting result as it suggests that the DCM provided in the solution (as shown in Figure reaction DMSO, reaction DCM) did not decrease the relative amount of byproduct to biotinylated-CHPA, but the overall use of biotin in the solution, perhaps through increasing the degree of intermediate formation during the first steps of the reaction.</p> | + | <p>MS analysis identified the control Biotin at the correct size as determined by the literature. The peak at 19 min. corresponds to the size of Biotin-CHPA plus a sodium ion (22.99 Da) which is found in high quanitites in the methanol used in electrospray which may be responsible for the higher peak size identified. Finally, the peak at 20 min. corresponded to the N-acylurea, the by-product of the reaction. This was an interesting result as it suggests that the DCM provided in the solution (as shown in Figure 7 reaction DMSO, reaction DCM) did not decrease the relative amount of byproduct to biotinylated-CHPA, but the overall use of biotin in the solution, perhaps through increasing the degree of intermediate formation during the first steps of the reaction.</p> |
<p>From the electrospray data, the peak at a retention time of 19 min. was determined to be the biotinylated product.</p> | <p>From the electrospray data, the peak at a retention time of 19 min. was determined to be the biotinylated product.</p> | ||
| + | <a name="biotin"></a> | ||
| + | <h2>Biotinylation of Other Naphthenic Acids</h2> | ||
| + | |||
| + | <p>Once cyclohexanepentanoic acid was identified to be biotinylated other compounds were selected and attempted to be biotinylated using the same procedure as above. Two other naphthenic acids, cyclohexanebutyric acid, and cyclohexanecarboxylic acid, were selected due to their availability, low cost, and differing side chains from CHPA. Their unique hydrophobic characters also allowed for our procedure to be tested against compounds which were slightly more hydrophilic than CHPA. Both of these compounds were incubated using the same reaction mixture as before and the solutions were run on HPLC, to analyze the success of these reactions.</p> | ||
| + | |||
| + | </html>[[Image:UofC_2011_CHBACHCA.png|thumb|625px|center|<b>Figure 9</b> Chemical structure of CHBA and CHCA, two other naphthenic acids which were conjugated to our biotinylated linker.]]<html> | ||
| + | </html>[[Image:UofC_2011_CHCACHBACHPA.png|thumb|625px|center|<b>Figure 10</b> HPLC analysis of CHBA, CHCA, and CHPA, comparing their unique peaks. CHBA and CHCA elute off of the column with a lower retention time than CHPA due to CHPA's hydrophobicity. All solutions were incubated overnight in the solvent solution described above.]]<html> | ||
| + | |||
| + | <p>Interestingly, the peaks identified by both compounds were very similar in layout to those identified for CHPA, suggesting that the compounds were being biotinylated and producing the same expected side reaction products. Both reactions additionally showed a reduction in retention time for all peaks which was expected. CHBA and CHCA were also analyzed alone, to identify their corresponding peaks throughout the time course.</p> | ||
| + | |||
| + | </html>[[Image:UofC_2011_PureCHCACHBA.png|thumb|625px|center|<b>Figure 11</b> HPLC analysis of pure CHCA (top) and CHBA (bottom) loaded at 50 ug. Each peak is denoted by a red arrow. Non-specific peaks in the CHCA run were a result of a previous run from a CHPA reaction solution which had been retained on the column. These peaks were not observed in any follow up runs using CHCA in reactions.]]<html> | ||
| + | |||
| + | |||
| + | <p>CHCA was found to have a retention time of approximately 12.5 minutes and CHBA, a retention time of <b>24</b> min. These corresponded to peaks in the reaction solution similar to that of CHPA. An advantage of using less hydrophobic compounds such as CHCA is that it allows for a decrease in the concentration of DMSO to be used in solution, making it easier to handle these biotinylated compounds for downstream aqueous applications. <b>Biotinylation of these three compounds, CHPA, CHBA, and CHCA identifies a proof of concept that our biotinylation strategy is effective at conjugating biotin-linkers to a variety of hydrophobic compounds.</b></p> | ||
| + | |||
| + | <h2>Biotinylated-Hydrophobic Small Molecules and Streptavidin Coated Beads</h2> | ||
| + | |||
| + | <p>With an optimized protocol for the biotinylation of small hydrophobic molecules, our team turned to identifying a method by which these conjugates could be efficiently taken up by streptavidin coated beads. Streptavidin and avidin are known to tightly interact with biotin, so much so that this interaction is considered one of the strongest non-covalent interactions known to man. The interaction is resistant to detergents, heat, alkalinity, reduction/oxidation, as well as a variety of other stresses making it a useful platform for use with hydrophobic small molecules. However, it was important that our team identify a method by which we could test the binding of our beads to our biotinylated naphthenic acids under non-polar solvent conditions. Commercially available streptavidin coated magnetic beads from Solulink were used to test this interaction. 100μg of beads were mixed with biotinylated naphthenic acids end-over-end for 30 min. at room temperature in a variety of solvent conditions and HPLC analysis was used to identify a change in peak height suggesting the beads were binding to the biotinylated compound (see Figure 12). </p> | ||
| + | |||
| + | </html>[[Image:UofC_2011_SBeadsData.png|thumb|625px|center|<b>Figure 12</b> HPLC analysis of 10 nmol biotin-linker incubated with and without 500 ug Solulink streptavidin coated magnetic beads end-over-end 30 min. room temperature. Samples were dried down using a vacufuge to remove solvent and resuspended in dH2O. Run's labelled with "Beads" were incubated, while those without this title had water added to the solution. All solutions were made to a 300 uL volume. Solvents were as follow: <i>+ve Control</i> - Water, <i>-ve Control</i> - 9% DCM/ 18% ddH2O / 73% DMSO, <i>No DCM</i> - 27% ddH2O / 73% DMSO, <i>1/2DMSO/DCM</i> - 4.5% DCM/ 59% ddH2O / 36.5% DMSO, <i>1/2 DMSO No DCM</i> - 63.5% ddH2O / 36.5% DMSO. Peaks were analyzed based on their height and width qualitatively.]]<html> | ||
| + | |||
| + | <p>Unfortunately, no significant shift in peak size was identified in peak height using the Solulink beads, even with the biotinylated linker alone dissolved in water. A distinct difference was noticed when beads were incubated with the 1/2DMSO No DCM runs, however, due to the lack of binding in water alone, further optimization was required. With these concerns a new bead platform would be required to increase the efficiency of this binding. Moving on, we attempted to optimize our biotinylated naphthenic acids using DynaBeads supplied by Invitrogen. Through use of their sample kit we were able to identify the relative amount of binding to the beads of our biotinylated linker alone in water using HPLC. | ||
| + | |||
| + | </html>[[Image:UofC_2011_IBeads.png|thumb|625px|center|<b>Figure 13</b> HPLC analysis of 10 nmol biotin-linker incubated with and without 100 uL Invitrogen streptavidin coated magnetic beads end-over-end 30 min. room temperature. Beads were incubated in 1x PBS Buffer pH 8.3. High pH was due to downstream application in order to keep hydrophobic compounds such as CHPA into solution allowing for binding to the beads. Four types of beads were tested and quantitated against a biotin control incubated without beads. All solutions were dried down and resuspended in water and 1/12th of the solution was injected onto the column. After quantification relative to the control, the 270 beads were identified to be the strongest binder of the biotinylated linker.]]<html> | ||
| + | |||
| + | <p>After quantification against four different kinds of Invitrogen DynaBeads, 270, was determined to be the best binder of the biotin-linker alone. 1xPBS pH 8.3 was used to test the beads as it was previously noted that numerous highly hydrophobic molecules (such as CHPA in its biotinylated form) seemed to readily stay in solution at the pH. This assay confirmed that the high pH of the PBS buffer did not comproomise the binding of biotin to streptavidin. This quantification identified the 270 streptavidin magnetic beads to be the best at binding this linker piece and the best type of bead system to use for downstream analysis with our buffer system. DMSO and DCM were not tested in this assay when unlike in the Solulink beads assay because the supplier was able to provide data suggesting that concentrations of DMSO at least up to 8% would not be harmful to the beads (Please contact Invitrogen for this information). | ||
<h2>Immunoprecipitation and Future Directions</h2> | <h2>Immunoprecipitation and Future Directions</h2> | ||
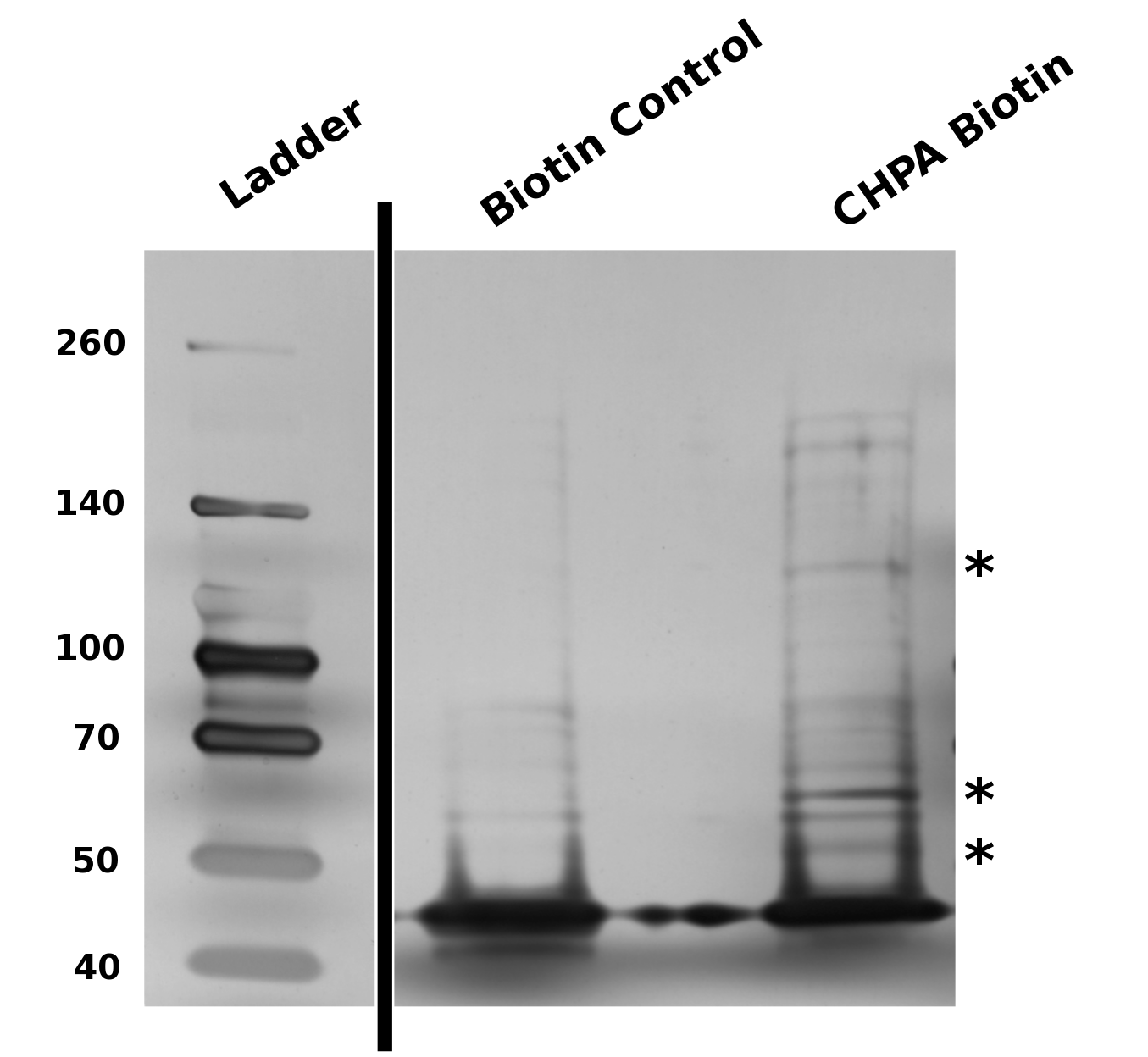
| - | <p>With the success of our biotinylation reaction, | + | <p>With the success of our biotinylation reaction, and the ability to determine a successful type of beads to perform our pull downs, we established a protocol by which we could perform a traditional IP for proteins which may bind to the cyclohexane ring of CHPA. This protocol can be found in the <a href="https://2011.igem.org/Team:Calgary/Notebook/Protocols/Process22">Protocols</a> section of the notebook in the menu. This procedure allowed us to successfully pull down protein interactors as seen in Figure 14. |
| - | < | + | </html>[[Image:UofC_2011_CHPAGel.png|thumb|625px|center|<b>Figure 14</b> BioRad 4-20% Bis-Tris gradient gel analysis of potential CHPA interacting proteins. Stars highlight enriched bands found in the CHPA elution but not identified in the Biotin control. For a step by step protocol of this procedure please see the protocols section.]]<html> |
| + | |||
| + | |||
| + | This pull-down suggests that our biotinylation scheme is capable of selectively pulling down protein elements specific to the conjugated element on the biotin linker. Our protocol is capable of enriching these interactions and concentrating them onto a gradient gel. The ability to pull down protein elements was the task which we chose to focus on as an initial proof of concept of this technique, however, one could adapt this protocol for the "fishing" of RNA, DNA (as in the example of chIP-SEQ), or other elements.</p> | ||
| + | |||
| + | <p>With isolated proteins, the University of Calgary team plans to determine their identity through mass spectrometry, and thus identify a potential naphthenic acid sensitive promoter. Additionally, by developing a larger consortium of biotinylated hydrophobic molecules, our team can identify unique responsive elements which may aid in the development of our biosensor. The biotinylation and immuno-based precipitation of small hydrophobic molecules is a novel approach to identification of protein binding partners in the literature and our team aims to optimize this further in the future</p> | ||
| - | |||
<h2>Conclusions</h2> | <h2>Conclusions</h2> | ||
| - | <p>A novel system for biotinylating cyclohexanepentanoic acid as well as other model naphthenic acids has been | + | <p>A novel system for biotinylating cyclohexanepentanoic acid as well as other model naphthenic acids has been developed and verified using HPLC and MS analysis. While further optimization of the protocols is required before performing IP based experiments, this technique provides the first steps toward a novel system to detect and isolate proteins involved in the naphthenic acid detection pathways of <i>Pseudomonas</i> and <i>Dunaliella</i>.</p> |
| + | |||
| + | <style> | ||
| + | #titlebar{margin-top: -6px;} | ||
| + | #pagetitle{padding-top: 17px;} | ||
| + | #bodycontainer{padding-top: 6px;} | ||
| + | </style> | ||
</html> | </html> | ||
}} | }} | ||
Latest revision as of 03:42, 29 October 2011









Novel Application of Biotinylation


Our team set out to identify a novel responsive element capable of detecting and quantifying naphthenic acids (NAs) in solution. While numerous studies have begun to identify species of bacteria which can survive and/or degrade NAs, the pathways of degradation have not yet been characterized. Therefore this goal required our team to design and implement novel methods to isolate the cellular systems of tailing ponds biological organisms which detect and potentially degrade naphthenic acids. We developed a naphthenic acid biotinylation/immunoprecipitation technique which allows for identification of novel protein complexes which interact and may potentially detect naphthenic acids. Our team developed an optimized protocol for the biotinylation of cyclohexanepentanoic acid (CHPA), cyclohexanebutyric acid (CHBA), and cyclohexanecarboxylic acid (CHCA), and a protocol for the immunoprecipitation of Pseudomonas lysates using commercially available streptavidin magnetic beads.
In addition, our team developed an in silico approach to isolating proteins which may be involved in degrading naphthenic acids using two specific species of Pseudomonas previously reported to degrade NAs in co-culture. Through use of various bioinformatic databases, a list of potential naphthenic acid degrading proteins was identified. Of these, a set were tested using reverse transcriptase polymerase chain reaction (RT-PCR) to identify a change in expression when incubated with naphthenic acids. A novel Enoyl-Coa-hydratase was identified and may be a responsive element to naphthenic acids.
Click here for more information about our Bioinformatic search.
Sensory Element Fishing: Designing A Novel Biotinylation-Immunprecipitation System For Isolation of Naphthenic Acid Interactors
What Are We Looking For?
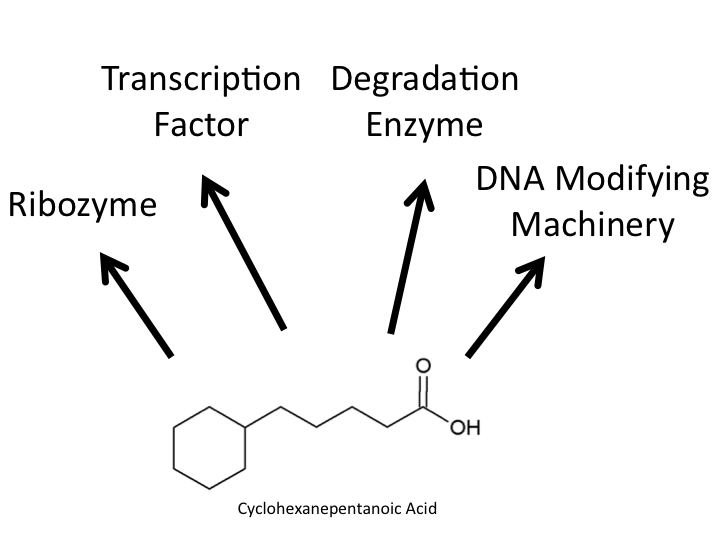
Due to the complexity of biological systems, our team focused our efforts on identifying a system for identification of proteins or complexes which bind DNA. Transcription factors, which bind to specific elements of a gene known as the promoter, have been well characterized in other metabolic systems to respond to metabolites in the cell and change the level of transcription of particular metabolic response genes. A common example of one of these systems is the Lac operon which has been harnessed as a selection system already in the registry. Identifying a system such as this for naphthenic acids, if one exists, would allow for a specific response to be generated to many different kinds of naphthenic acids. Our hypothesis requires that the organisms we use respond specifically to naphthenic acids and result in specific upregulation of metabolic genes with little background effect in the cell. Additionally, numerous protein elements have been shown to bind and modify DNA, resulting in downstream changes to protein expression. Examples of this are histone modifying enzymes in Eukaryotes which have opened up a new realm of research known as epigenetics. These systems can be complicated and often do not directly recognize metabolic chemical compounds. Furthermore, it is possible that naphthenic acids may even interact with specific RNA responsive elements known as riboswitches, which may be involved in the modification of protein expression. While the identification of any of these systems were of specific interest to us to identify a detection system for naphthenic acids, we wanted our approach to be broad enough to idenitfy any components interacting with naphthenic acids.
Naphthenic Acid Degrading Organisms To Be Used
Due to the diversity and ambiguity of microbial species in the tailings ponds, we chose to focus our efforts for our promoter search on two species of Pseudomonas and a species of microalgae Dunaliella tertiolecta (please see our chassis section for more information on these species). All of these organisms have been shown to degrade naphthenic acids, however little work has been done in characterizing the degradation pathways of both species.
Method Design
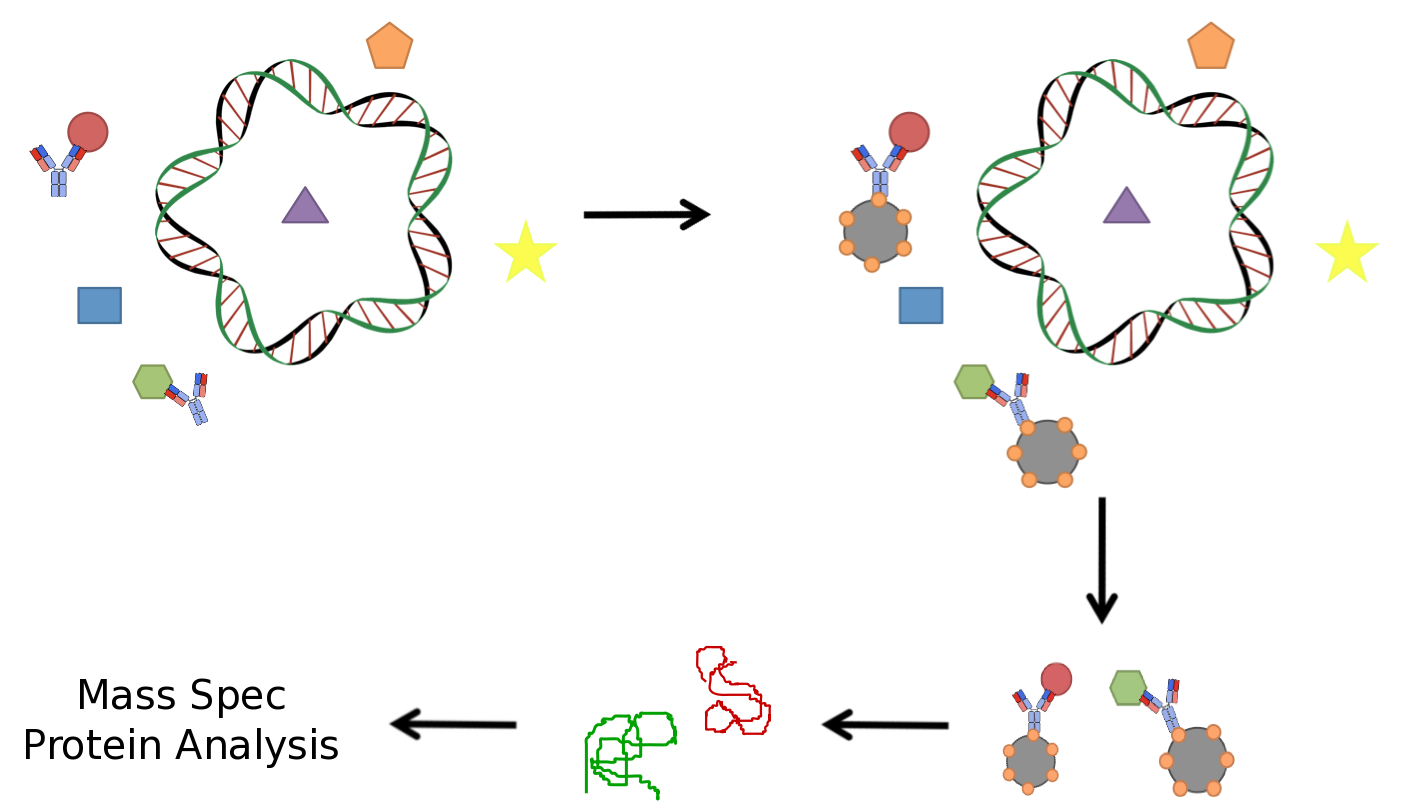
As we plan to screen both bacterial and microalgae species, it was vital that our screening technique be robust and efficient in a variety of cellular systems. It was also important that it selectively targeted protein elements which were specific for naphthenic acids and did not result in unwanted background. A common technique used in similar applications including protein biochemistry is immunoprecipitation (IP) for the identification of protein interactors against a particular bait protein. An antibody is incubated with the lysate of the organism of interest. The target of this antibody and its interacting proteins can be isolated using beads that bind to a conserved region opposite the antigen binding domain. Isolation of the target protein along with its interactors allows for their identification through mass spectrometry (MS), a routinely used technique to identify proteins. This technique has been well established and is used routinely in a variety of applications.
In a similar manner we designed a protocol to use naphthenic acids in an immunoprecipitation. This however, required that we develop a system to conjugate our naphthenic acids to a bead platform similar to the commercially available antibodies used in traditional IP. To accomplish this, we attempted to react the conserved carboxylic acid of the naphthenic acid cyclohexanepentanoic acid (CHPA) (see overview section) with a commercially available biotin-amine derivative. By reacting these compounds in the presence of EDC (1-ethyl-3-(3-dimethylaminopropyl)carbodiimide) the carboxylic acid and primary amine can produce an amide linkage under low pH conditions. Biotinylation is commonly performed with DNA and protein side chains using commercial kits designed for these applications. However, the biotinylation of small molecules is rare and techniques involving hydrophobic compounds such as CHPA are not found in the literature. This required that we generate a novel protocol for biotinylating CHPA along with other naphthenic acid compounds.
Once our naphthenic acid has become biotinylated, the biotin group will strongly interact with a protein known as streptavidin. The biotin/streptavidin interaction is one of the strongest binding interactions characterized in biology (Kd=1x10-15M) and is commonly cross-linked to magnetic beads allowing for our biotinylated naphthenic acid to be used in similar protocols as in immunoprecipitation. In theory, proteins which interact with the cyclic ring structure of cyclohexane pentanoic acid can be isolated using our biotinylated CHPA/streptavidin beads platform. Finally mass spectrometry can be used to determine the identity of these regulatory elements (See Figure 3).
Biotinylation
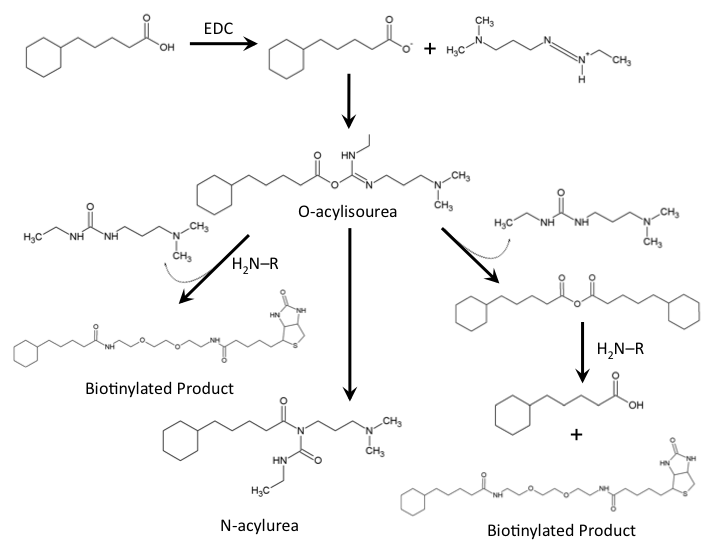
The biotinylation of molecules containing carboxylic acid groups follows a well characterized reaction mechanism. In the context of our biotinylation reagents (CHPA and Biotin-PEG-amine) the reaction mechanism can be seen in Figure 4. The cyclohexanepentanoic acid will react with the carbodiimide (EDC) to produce an O-acylisourea intermediate activating the carboxylic acid making it an excellent leaving group. This requires a low pH environment to deprotonate the carboxylic acid. From this intermediate the desired product can be produced via direct attack of the negatively charged carboxylic acid with the amine group of our biotinylated compound producing a urea based compound as a by-product. Further reaction of the O-acylisourea intermediate with cyclohexanepentanoic acid may result in an acid anhydride which then produces the biotinylated compound of interest. The main byproduct of the reaction involves the rearrangement of the O-acylisourea to a stable N-acylurea. Common solutions to reduce the reaction towards this product involves the use of low-dielectric constant solvents such as dichloromethane. Due to the hydrophobic nature of cyclohexanepentanoic acid, aqueous low pH solutions will cause precipitation of this reaction substrate, therefore, the use of other solvents such as dimethylsulfoxide need to be used. The final reaction solution used in all biotinylation experiments used 9% DCM/ 18% ddH2O / 73% DMSO. Typical reactions run overnight at room temperature. (Please see our protocols page for more information on the biotinylation experiment).
Biotinylation Confirmation
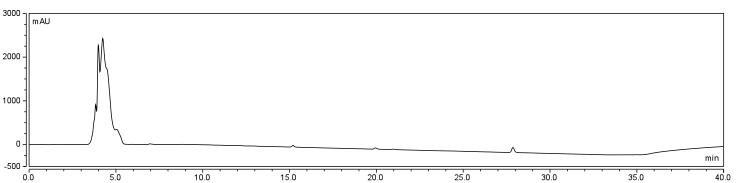
In order to confirm the biotinylation reaction was occurring, reverse phase High Performance Liquid Chromatography (HPLC) was used. A C18 analytical column (Dionex Acclaim 120) connected to a Dionex ICS-5000 series HPLC was adapted from a program which was based on complex naphthenic acid solution.
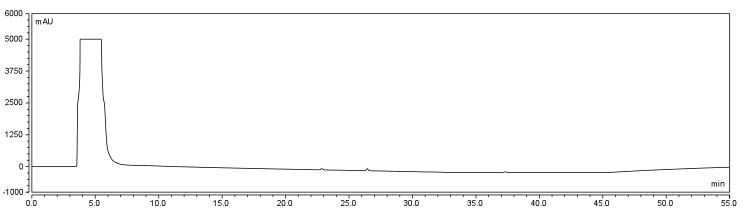
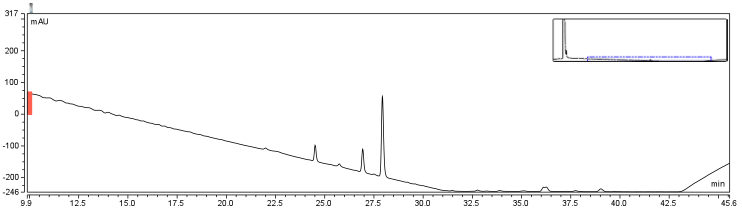
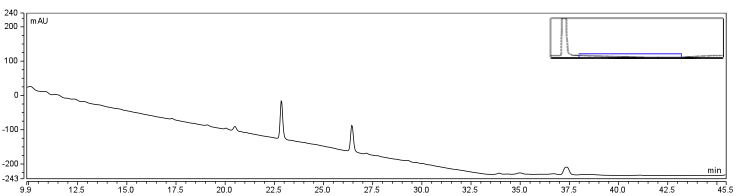
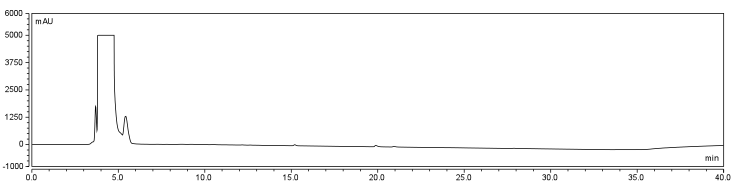
The HPLC conditions used for the analysis were as follows. A 60 min. program was designed with a mobile phase of 40% Acetonitrile / 60% 0.1% TFA Water with a gradient from 3-28 min. changing to 85% Acetonitrile/15% 0.1% TFA Water solution holding until 40 min. The final 5-20 min. of the program (runs later in the year were modulated such that there was only a 5 minute ramp back to starting conditions) ramped back to original gradient to clean the column. A 25 μL sample loop was used for all injections except for those used in purifying large amounts of biotinylated compound where a 500 μL sample loop was used. Samples were loaded onto the column at a flow rate of 0.6 mL/min at a temperature of 30 To establish a set of control conditions for this data analysis, DMSO and DCM were individually injected onto the column and a read out at 210 nm on a UV/VIS spectrophotometer over a 60 min. Due to the large peak produced by both of these solvents, it was thought that identification of potential aqueous solutions which elute between 3-7 min. of the program may be masked by the large peak produced during elution of DMSO or DCM. Therefore to examine the peaks for the reactants of the solution (biotin, and EDAC) were examined in water solutions. CHPA was diluted in 80% DMSO. Biotin, EDAC, and CHPA were loaded onto the column at 25 uL of 100 mM, 500 mM, and 2mg/mL stock solutions Due to the retention time of biotin and EDAC being within the rentention time of DMSO, there presence was difficult to detect during the biotinylation reaction. A reaction was set up in 80% DMSO/ 20% ddH2O to test the efficiency of reaction. A control and reaction was set up as written in the protocols section of the wiki, and incubated O/N at room temperature. Four peaks were observed in the reaction dataset which was not observed in the control with retention times of 15 min. 19 min. 20 min. and 28 min. The 28 min. peak was found to correspond to the CHPA control. Therefore the remaining three unique peaks were determined to show that the reaction was progressing. Independent HPLC analysis showed that after 24 hours the reaction had gone to completion and there was no difference in peak sizes observed (data not shown).
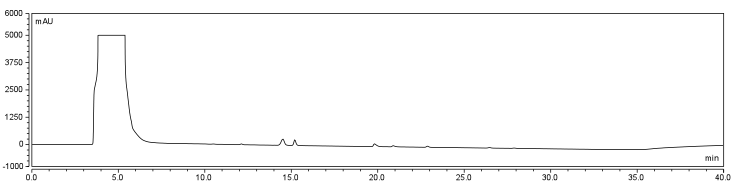
In order to further optimize this reaction, the reaction was repeated with and without the use of 9% DCM, in the same conditions as before. Comparison of the two datasets shows an increase in the three reaction peaks with relatively no change in the CHPA peak, suggesting that dichloromethane’s low dielectric constant increases the relative rate of reaction. Finally, to confirm the identity of our biotinylated product, peak fractions were collected for a potential biotin peak (~4 min.), the peak at 19 min. and the second peak at 20 min. The expected masses of biotin control and the expected products are as follows:
Samples were collected from the HPLC in 30 sec. fractions of 0.3mL and dried down in a vacufuge at 45C. Samples were submitted to the University of Calgary Department of Chemistry MS Facility for electrospray analysis.
MS analysis identified the control Biotin at the correct size as determined by the literature. The peak at 19 min. corresponds to the size of Biotin-CHPA plus a sodium ion (22.99 Da) which is found in high quanitites in the methanol used in electrospray which may be responsible for the higher peak size identified. Finally, the peak at 20 min. corresponded to the N-acylurea, the by-product of the reaction. This was an interesting result as it suggests that the DCM provided in the solution (as shown in Figure 7 reaction DMSO, reaction DCM) did not decrease the relative amount of byproduct to biotinylated-CHPA, but the overall use of biotin in the solution, perhaps through increasing the degree of intermediate formation during the first steps of the reaction.
From the electrospray data, the peak at a retention time of 19 min. was determined to be the biotinylated product.
Biotinylation of Other Naphthenic Acids
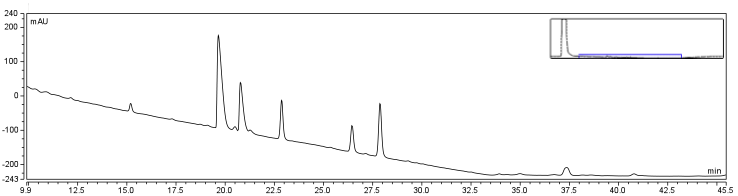
Once cyclohexanepentanoic acid was identified to be biotinylated other compounds were selected and attempted to be biotinylated using the same procedure as above. Two other naphthenic acids, cyclohexanebutyric acid, and cyclohexanecarboxylic acid, were selected due to their availability, low cost, and differing side chains from CHPA. Their unique hydrophobic characters also allowed for our procedure to be tested against compounds which were slightly more hydrophilic than CHPA. Both of these compounds were incubated using the same reaction mixture as before and the solutions were run on HPLC, to analyze the success of these reactions.
Interestingly, the peaks identified by both compounds were very similar in layout to those identified for CHPA, suggesting that the compounds were being biotinylated and producing the same expected side reaction products. Both reactions additionally showed a reduction in retention time for all peaks which was expected. CHBA and CHCA were also analyzed alone, to identify their corresponding peaks throughout the time course.

CHCA was found to have a retention time of approximately 12.5 minutes and CHBA, a retention time of 24 min. These corresponded to peaks in the reaction solution similar to that of CHPA. An advantage of using less hydrophobic compounds such as CHCA is that it allows for a decrease in the concentration of DMSO to be used in solution, making it easier to handle these biotinylated compounds for downstream aqueous applications. Biotinylation of these three compounds, CHPA, CHBA, and CHCA identifies a proof of concept that our biotinylation strategy is effective at conjugating biotin-linkers to a variety of hydrophobic compounds.
Biotinylated-Hydrophobic Small Molecules and Streptavidin Coated Beads
With an optimized protocol for the biotinylation of small hydrophobic molecules, our team turned to identifying a method by which these conjugates could be efficiently taken up by streptavidin coated beads. Streptavidin and avidin are known to tightly interact with biotin, so much so that this interaction is considered one of the strongest non-covalent interactions known to man. The interaction is resistant to detergents, heat, alkalinity, reduction/oxidation, as well as a variety of other stresses making it a useful platform for use with hydrophobic small molecules. However, it was important that our team identify a method by which we could test the binding of our beads to our biotinylated naphthenic acids under non-polar solvent conditions. Commercially available streptavidin coated magnetic beads from Solulink were used to test this interaction. 100μg of beads were mixed with biotinylated naphthenic acids end-over-end for 30 min. at room temperature in a variety of solvent conditions and HPLC analysis was used to identify a change in peak height suggesting the beads were binding to the biotinylated compound (see Figure 12).
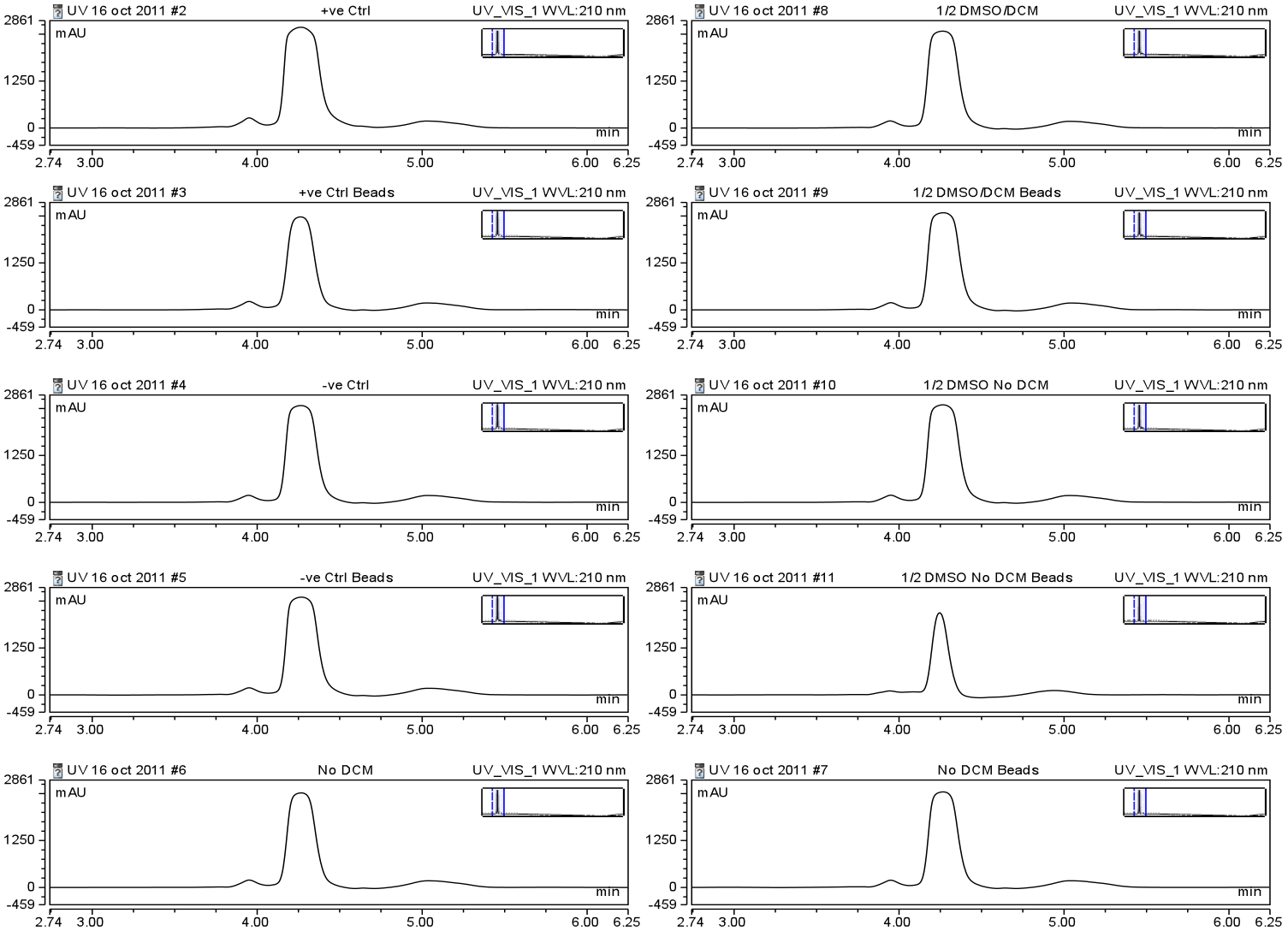
Unfortunately, no significant shift in peak size was identified in peak height using the Solulink beads, even with the biotinylated linker alone dissolved in water. A distinct difference was noticed when beads were incubated with the 1/2DMSO No DCM runs, however, due to the lack of binding in water alone, further optimization was required. With these concerns a new bead platform would be required to increase the efficiency of this binding. Moving on, we attempted to optimize our biotinylated naphthenic acids using DynaBeads supplied by Invitrogen. Through use of their sample kit we were able to identify the relative amount of binding to the beads of our biotinylated linker alone in water using HPLC.
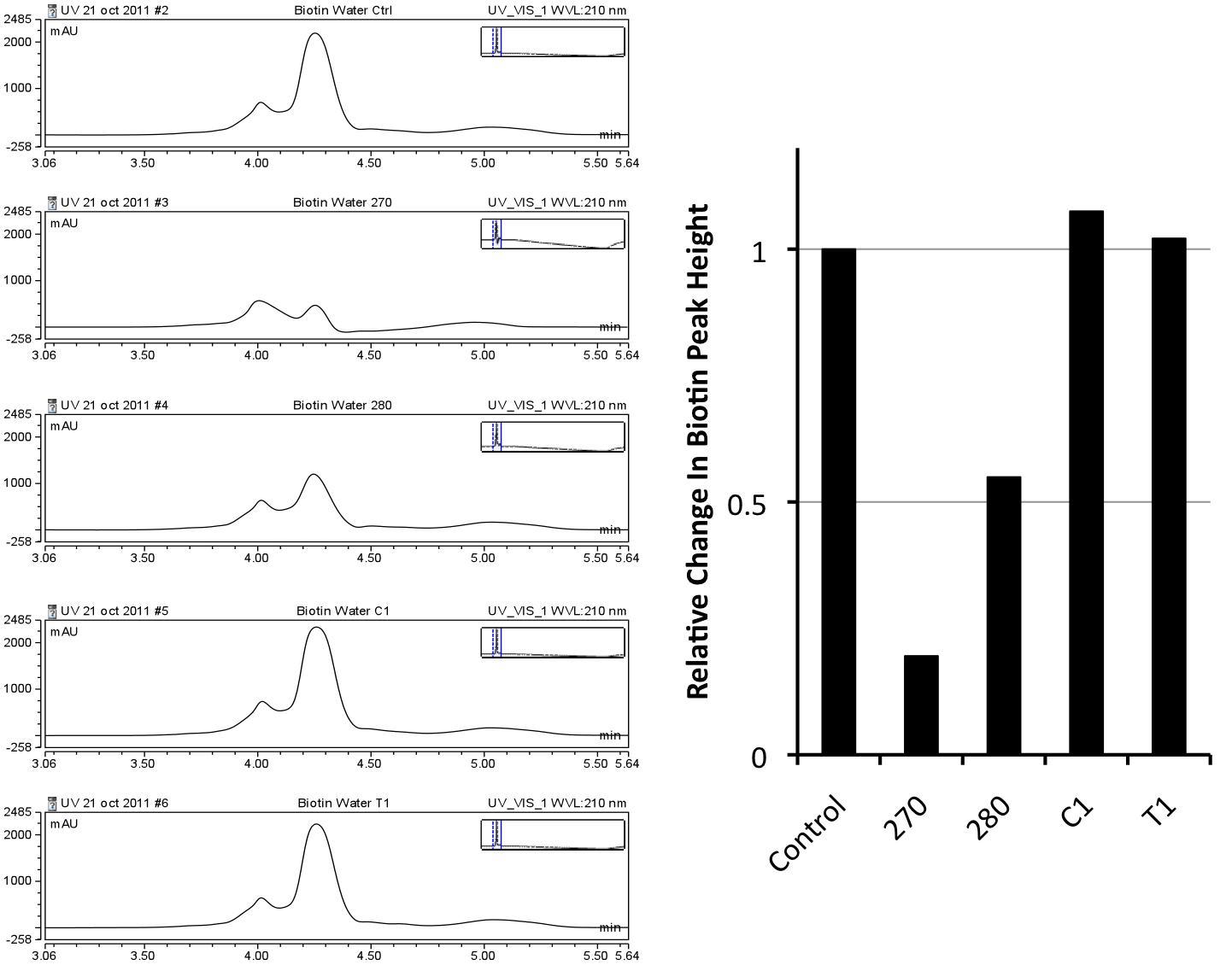
After quantification against four different kinds of Invitrogen DynaBeads, 270, was determined to be the best binder of the biotin-linker alone. 1xPBS pH 8.3 was used to test the beads as it was previously noted that numerous highly hydrophobic molecules (such as CHPA in its biotinylated form) seemed to readily stay in solution at the pH. This assay confirmed that the high pH of the PBS buffer did not comproomise the binding of biotin to streptavidin. This quantification identified the 270 streptavidin magnetic beads to be the best at binding this linker piece and the best type of bead system to use for downstream analysis with our buffer system. DMSO and DCM were not tested in this assay when unlike in the Solulink beads assay because the supplier was able to provide data suggesting that concentrations of DMSO at least up to 8% would not be harmful to the beads (Please contact Invitrogen for this information).
Immunoprecipitation and Future Directions
With the success of our biotinylation reaction, and the ability to determine a successful type of beads to perform our pull downs, we established a protocol by which we could perform a traditional IP for proteins which may bind to the cyclohexane ring of CHPA. This protocol can be found in the Protocols section of the notebook in the menu. This procedure allowed us to successfully pull down protein interactors as seen in Figure 14.
With isolated proteins, the University of Calgary team plans to determine their identity through mass spectrometry, and thus identify a potential naphthenic acid sensitive promoter. Additionally, by developing a larger consortium of biotinylated hydrophobic molecules, our team can identify unique responsive elements which may aid in the development of our biosensor. The biotinylation and immuno-based precipitation of small hydrophobic molecules is a novel approach to identification of protein binding partners in the literature and our team aims to optimize this further in the future
Conclusions
A novel system for biotinylating cyclohexanepentanoic acid as well as other model naphthenic acids has been developed and verified using HPLC and MS analysis. While further optimization of the protocols is required before performing IP based experiments, this technique provides the first steps toward a novel system to detect and isolate proteins involved in the naphthenic acid detection pathways of Pseudomonas and Dunaliella.

 "
"