Team:WashU/Modeling
From 2011.igem.org
(→Modeling) |
J.herrmann (Talk | contribs) (→Modeling) |
||
| (24 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{WashUHeader2011}} | {{WashUHeader2011}} | ||
| - | + | ==Modeling== | |
| - | [[File:MasterPlan1.jpg|thumb| | + | [[File:Pathway1.jpg|thumb|left|upright=3]] |
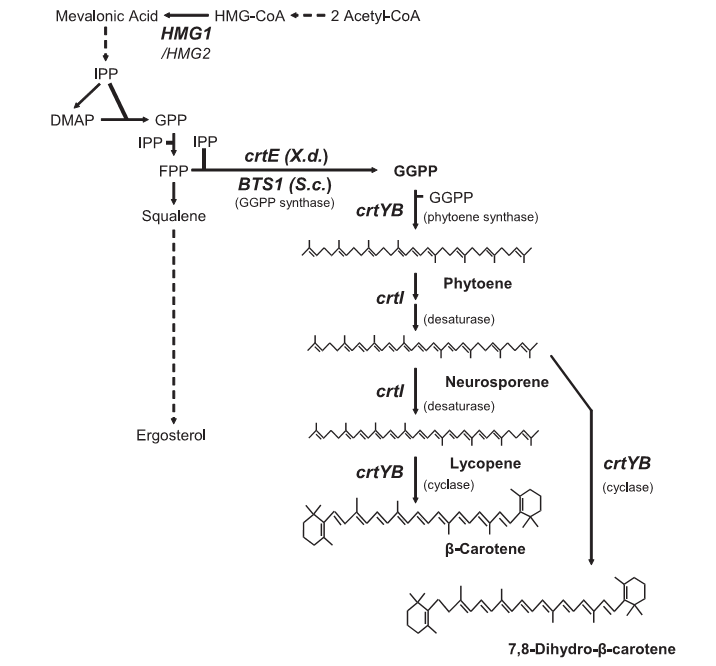
| + | The pathway for producing beta-carotene as shown in "''High-Level Production of Beta-Carotene in Saccharomyces cerevisiae by Successive Transformation with Carotenogenic Genes from Xanthophyllomyces dendrorhous''" by Rene Verwaal et. Al. Yeast naturally produce GGPP but no other intermediates in beta-carotene pathway, crtYB and crtI are needed to to complete the pathway for beta-carotene production. Notice crtYB codes for two proteins needed in the pathway. CrtE further improves beta-carotene production as it aides in the production of the starting product, GGPP. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | [[File:Pathway2.jpg|thumb|left|upright=3]] | ||
| + | |||
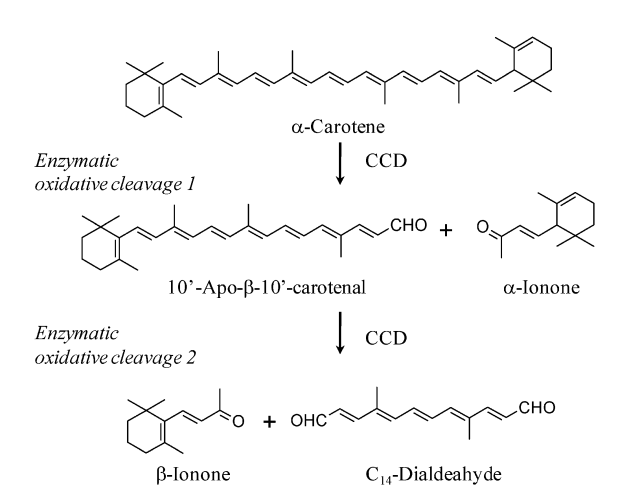
| + | The pathway for producing beta-ionone from beta-carotene as shown in "''Functional Characterization of a Carotenoid Cleavage Dioxygenase 1 and Its Relation to the Carotenoid Accumulation and Volatile Emission During the Floral | ||
| + | Development of Osmanthus Fragrans Lour''" by Susanne Baldermann et. Al. This process only involves the expression of one gene, CCD, which codes for enzymatic oxidative cleavage 1 and 2, which cleaves the ends of beta-carotene to produce two beta-ionone molecules. | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | == '''Experimental Plan''' == | ||
| + | |||
| + | [[File:MasterPlan1.jpg|thumb|left|upright=3]] | ||
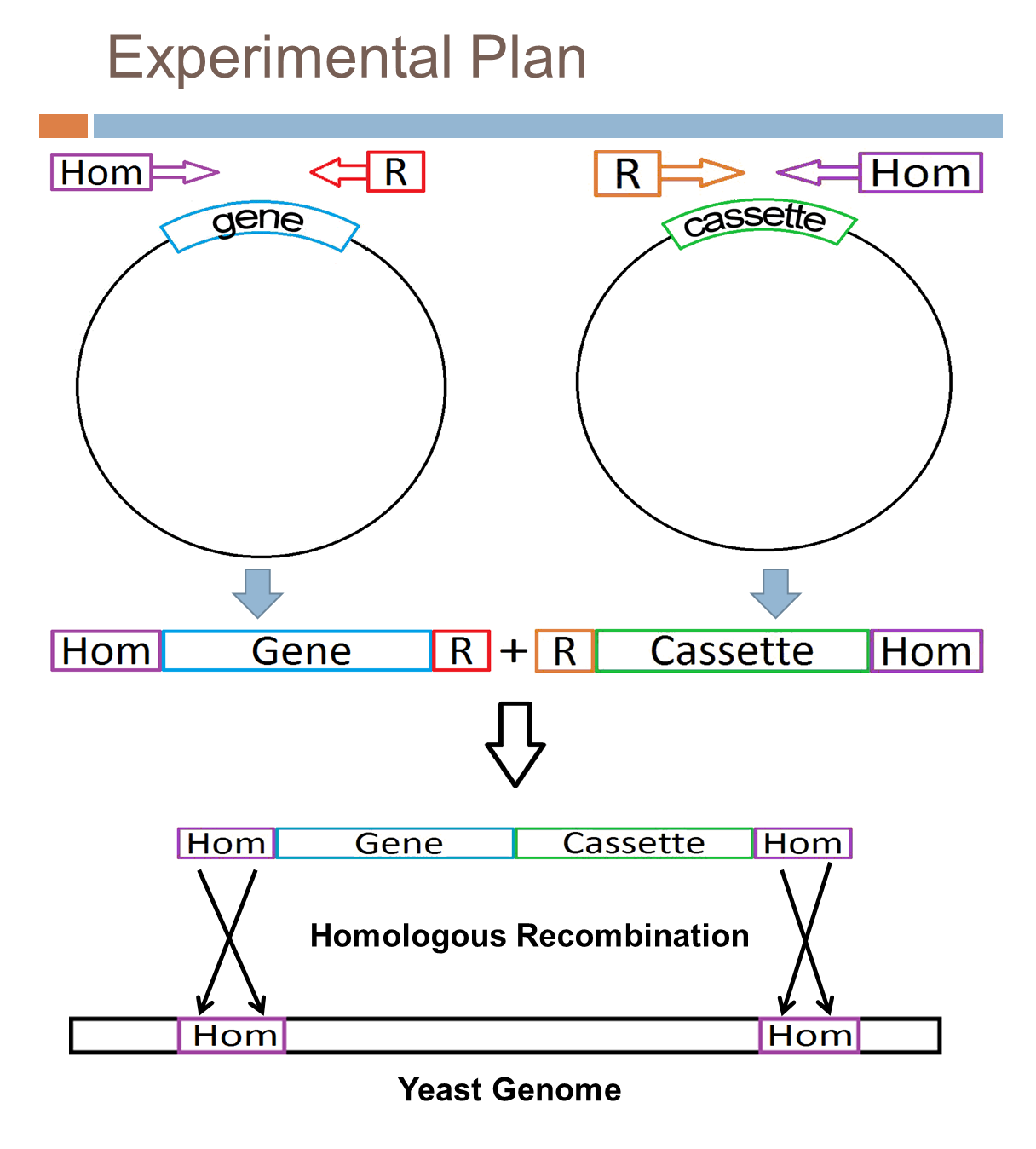
| + | The goal of the project is to express the four previously mentioned genes, crtI, crtYB, crtE, and CCD1, needed for the beta-carotene and beta-ionone synthesis pathway in yeast. | ||
| + | *Each of the four genes is first synthesized. | ||
| + | *Then a restriction site and sections homologous to a suitable section of the yeast genome are added to the ends of the genes. | ||
| + | *In parallel, a cassette containing a gene that allows for separation of transformed yeast from untransformed (such as ampicillin resistance) is modified in the same way. (Restriction enzymes: Avr2 and Spe1) | ||
| + | |||
| + | |||
| + | *The gene and cassette are then ligated together using the restriction sites. | ||
| + | *The gene/cassette DNA is then introduced to yeast cells. | ||
| + | |||
| + | |||
| + | *Homologous recombination occurs between homologous ends of the gene/cassette and yeast gene naturally, albeit infrequently. | ||
| + | |||
| + | *The gene/cassette is now integrated into the genome and therefore expressed by yeast. | ||
Latest revision as of 07:20, 28 September 2011
Modeling
The pathway for producing beta-carotene as shown in "High-Level Production of Beta-Carotene in Saccharomyces cerevisiae by Successive Transformation with Carotenogenic Genes from Xanthophyllomyces dendrorhous" by Rene Verwaal et. Al. Yeast naturally produce GGPP but no other intermediates in beta-carotene pathway, crtYB and crtI are needed to to complete the pathway for beta-carotene production. Notice crtYB codes for two proteins needed in the pathway. CrtE further improves beta-carotene production as it aides in the production of the starting product, GGPP.
The pathway for producing beta-ionone from beta-carotene as shown in "Functional Characterization of a Carotenoid Cleavage Dioxygenase 1 and Its Relation to the Carotenoid Accumulation and Volatile Emission During the Floral Development of Osmanthus Fragrans Lour" by Susanne Baldermann et. Al. This process only involves the expression of one gene, CCD, which codes for enzymatic oxidative cleavage 1 and 2, which cleaves the ends of beta-carotene to produce two beta-ionone molecules.
Experimental Plan
The goal of the project is to express the four previously mentioned genes, crtI, crtYB, crtE, and CCD1, needed for the beta-carotene and beta-ionone synthesis pathway in yeast.
- Each of the four genes is first synthesized.
- Then a restriction site and sections homologous to a suitable section of the yeast genome are added to the ends of the genes.
- In parallel, a cassette containing a gene that allows for separation of transformed yeast from untransformed (such as ampicillin resistance) is modified in the same way. (Restriction enzymes: Avr2 and Spe1)
- The gene and cassette are then ligated together using the restriction sites.
- The gene/cassette DNA is then introduced to yeast cells.
- Homologous recombination occurs between homologous ends of the gene/cassette and yeast gene naturally, albeit infrequently.
- The gene/cassette is now integrated into the genome and therefore expressed by yeast.
 "
"