Team:BU Wellesley Software/Project Overview
From 2011.igem.org
m |
|||
| (18 intermediate revisions not shown) | |||
| Line 38: | Line 38: | ||
text-transform: uppercase; | text-transform: uppercase; | ||
text-decoration: none; | text-decoration: none; | ||
| - | text-align: | + | text-align: center; |
color: #3d3f3c; | color: #3d3f3c; | ||
font-size: 32pt; | font-size: 32pt; | ||
| Line 89: | Line 89: | ||
<br> | <br> | ||
<h6>Project Overview</h6> | <h6>Project Overview</h6> | ||
| + | <br><br> | ||
| + | <center> | ||
| + | <a href="https://2011.igem.org/Team:BU_Wellesley_Software/Clotho">Clotho</a> | <a href="https://2011.igem.org/Team:BU_Wellesley_Software/G-nomeSurferPro">G-nome Surfer Pro</a> | <a href="https://2011.igem.org/Team:BU_Wellesley_Software/OptimusPrimer">Optimus Primer</a> | <a href="https://2011.igem.org/Team:BU_Wellesley_Software/Trumpet">Trumpet</a> | <a href="https://2011.igem.org/Team:BU_Wellesley_Software/Puppetshow">Puppetshow</a> | <a href="https://2011.igem.org/Team:BU_Wellesley_Software/eLabNotebook">eLabNotebook</a> | <a href="https://2011.igem.org/Team:BU_Wellesley_Software/Wet_Lab">Wet Lab</a> | <a href="https://2011.igem.org/Team:BU_Wellesley_Software/Downloads_and_Tutorials">Downloads and Tutorials</a> | ||
| + | </center> | ||
<p> | <p> | ||
| + | <br><br> | ||
<p><p> | <p><p> | ||
| + | <center> | ||
<div id="project_wheel"> | <div id="project_wheel"> | ||
<img name="wheel" src="http://cs.wellesley.edu/~hcilab/iGEM_wiki/images/ProjectLogos/wheel.png" width="700px" alt="wheel" usemap="#wheelmap" > | <img name="wheel" src="http://cs.wellesley.edu/~hcilab/iGEM_wiki/images/ProjectLogos/wheel.png" width="700px" alt="wheel" usemap="#wheelmap" > | ||
| Line 117: | Line 123: | ||
</map> | </map> | ||
</div> | </div> | ||
| + | </center> | ||
<p> | <p> | ||
<div class="project_abstract"> | <div class="project_abstract"> | ||
| - | <p> | + | <!--<p> |
| - | We present tools which facilitate the research, design, and fabrication of biological constructs. Our workflow comprises | + | We present tools which facilitate the research, design, and fabrication of biological constructs. Our workflow comprises <a href="https://2011.igem.org/Team:BU_Wellesley_Software/G-nomeSurferPro">G-nome Surfer Pro</a> for research, <a href="https://2011.igem.org/Team:BU_Wellesley_Software/Trumpet">Trumpet</a> and <a href="https://2011.igem.org/Team:BU_Wellesley_Software/OptimusPrimer">Optimus Primer</a> for design, and <a href="https://2011.igem.org/Team:BU_Wellesley_Software/Puppetshow">Puppetshow</a> and <a href="https://2011.igem.org/Team:BU_Wellesley_Software/eLabNotebook">eLabNotebook</a> for the construction of those designs. <a href="https://2011.igem.org/Team:BU_Wellesley_Software/G-nomeSurferPro">G-nome Surfer Pro</a> promotes collaborative |
research by allowing users to browse genes, Parts, and other DNA along with their associated | research by allowing users to browse genes, Parts, and other DNA along with their associated | ||
| - | literature on a table-top surface. Using Optimus Primer, primers can be designed for the selected | + | literature on a table-top surface. Using <a href="https://2011.igem.org/Team:BU_Wellesley_Software/OptimusPrimer">Optimus Primer</a>, primers can be designed for the selected |
| - | Parts. Trumpet generates permutable constructs from these Parts by interleaving invertase sites | + | Parts. <a href="https://2011.igem.org/Team:BU_Wellesley_Software/Trumpet">Trumpet</a> generates permutable constructs from these Parts by interleaving invertase sites |
among them. To assemble these permutable constructs, we present a Protocol Automation Stack | among them. To assemble these permutable constructs, we present a Protocol Automation Stack | ||
| - | comprising a high-level programming language called | + | comprising a high-level programming language called <a href="https://2011.igem.org/Team:BU_Wellesley_Software/Puppetshow">Puppetshow</a>, executable on a robot. For improving |
| - | manual protocol execution, we are developing an | + | manual protocol execution, we are developing an <a href="https://2011.igem.org/Team:BU_Wellesley_Software/eLabNotebook">eLabNotebook</a> that helps capture data, and |
schedule resources and lab activities. Our unique tools offer an end-to-end workflow that is | schedule resources and lab activities. Our unique tools offer an end-to-end workflow that is | ||
collaborative, includes wetlab automation and organization, and provides algorithms for designing | collaborative, includes wetlab automation and organization, and provides algorithms for designing | ||
configurable constructs. | configurable constructs. | ||
| + | </p>--> | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | <h1>Design Cycle</h1> | ||
| + | <p> | ||
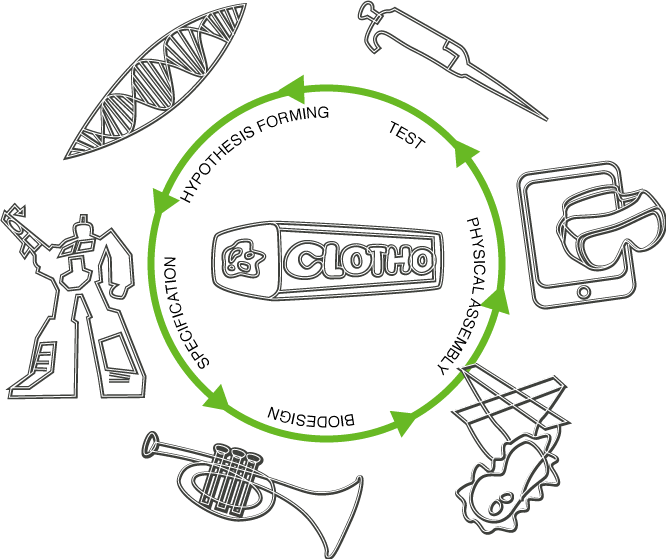
| + | Synthetic biological systems of the future will require that software design flows and laboratory experimentation be closely coupled into what is commonly referred to as the design-build-test cycle. Our project expands on this process by creating a collection of novel software tools. These software tools not only tackle challenging technical problems (automated genetic circuit design, liquid handling robot automation) but also introduce innovative approaches to the way in which users interact both with each other as well as with computers. In addition, we carry out wetlab experiments using our tools to validate our approach. We feel that all three areas (technical solutions, innovative interfaces, and wetlab validation) are crucial and allow our project to standout from traditional iGEM software projects. Our design cycle is organized around the following five areas: | ||
| + | |||
| + | <ol> | ||
| + | <li><b>Hypothesis forming</b> - In this stage, high level decisions are made regarding the system architecture, desired functionality, relevant background literature, and overall vision of the project and system. | ||
| + | <ul><li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/G-nomeSurferPro">G-nome Surfer Pro</a> promotes collaborative research by allowing users to browse genes, Parts, and other DNA along with their associated literature on a table-top surface. | ||
| + | </ul> | ||
| + | |||
| + | <li><b>Specification</b> - Here the required system components are formally captured and any constraints on the design are specified. | ||
| + | <ul><li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/OptimusPrimer">Optimus Primer</a> provides both a desktop as well as a table-top surface environment to generate primers for selected Parts from the hypothesis forming stage. | ||
| + | </ul> | ||
| + | |||
| + | <li><b>Design</b> - This is the process of transforming the specification into a collection of biological building blocks with sufficient information to physically assemble them in the laboratory environment. | ||
| + | <ul><li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Trumpet">Trumpet</a> generates permutable genetic circuit constructs from the Specification stage designs by interleaving invertase sites at strategic locations within genetic Devices. | ||
| + | </ul> | ||
| + | |||
| + | <li><b>Physical Assembly</b> - Assembling DNA constructs is a series of transformations which take DNA primitives and combine them into Composite Parts. Issues such as efficiency, design cost, and reliability should be taken into account using optimized algorithms and tools. | ||
| + | <ul><li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Puppetshow">Puppetshow</a> assembles these permutable constructs from the Design stage using a Protocol Automation Stack comprising a high-level programming language executable on a liquid handling robot. | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/eLabNotebook">eLabNotebook</a> helps capture data, and schedule resources and lab activities creating a more design automation friendly laboratory environment. | ||
| + | </ul> | ||
| + | |||
| + | <li><b>Test</b> - Finally, the assembled construct must be verified. In the event that it does not work, the design cycle can continue incorporating the knowledge gained at this stage to try to improve the overall process. | ||
| + | <ul><li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Wet_Lab">Wet Lab</a> experimentation tested our tools and methodology by beginning to create a framework for investigating transcription factor interactions in Tuberculosis. | ||
| + | </ul> | ||
| + | </ol> | ||
</p> | </p> | ||
| + | <p> | ||
| + | |||
| + | <h1>Integration</h1> | ||
| + | <p> | ||
| + | In order to integrate these areas fully together we built upon the <a href="https://2011.igem.org/Team:BU_Wellesley_Software/Clotho">Clotho</a> design framework. Clotho is a portable and extensible collection of open-source application software, a concise data model, and a core software development platform for synthetic biological engineering. Being open-source and easily extensible, Clotho is built to address the current and future needs of synthetic biologists. Clotho's data model captures the data associated with synthetic biological parts, devices, systems, and samples, in a unified relational database. Clotho's application software is modularized into "Apps" which all exchange biological data with a database through a uniform interface. Furthermore, Clotho's core platform introduces a concise and simple application programming interface (API) to developers who wish to contribute new applications to Clotho and create customized in-house tools. The API establishes a vocabulary at a level of abstraction familiar to synthetic biologists. </p> | ||
| + | |||
</div><br> | </div><br> | ||
</div><!--end bu-wellesley_wiki_content div--> | </div><!--end bu-wellesley_wiki_content div--> | ||
| + | |||
| + | <br> | ||
| + | For a more extended overview, check out our talk at the Americas Regional competition: | ||
| + | <br> | ||
| + | <br> | ||
| + | |||
| + | <center> | ||
| + | <iframe width="560" height="315" src="http://www.youtube.com/embed/xVfQyxbHEhU" frameborder="0" allowfullscreen </iframe> | ||
| + | </center> | ||
| + | |||
| + | |||
</body> | </body> | ||
</html> | </html> | ||
Latest revision as of 20:01, 18 October 2011
Project Overview
Design Cycle
Synthetic biological systems of the future will require that software design flows and laboratory experimentation be closely coupled into what is commonly referred to as the design-build-test cycle. Our project expands on this process by creating a collection of novel software tools. These software tools not only tackle challenging technical problems (automated genetic circuit design, liquid handling robot automation) but also introduce innovative approaches to the way in which users interact both with each other as well as with computers. In addition, we carry out wetlab experiments using our tools to validate our approach. We feel that all three areas (technical solutions, innovative interfaces, and wetlab validation) are crucial and allow our project to standout from traditional iGEM software projects. Our design cycle is organized around the following five areas:
- Hypothesis forming - In this stage, high level decisions are made regarding the system architecture, desired functionality, relevant background literature, and overall vision of the project and system.
- G-nome Surfer Pro promotes collaborative research by allowing users to browse genes, Parts, and other DNA along with their associated literature on a table-top surface.
- Specification - Here the required system components are formally captured and any constraints on the design are specified.
- Optimus Primer provides both a desktop as well as a table-top surface environment to generate primers for selected Parts from the hypothesis forming stage.
- Design - This is the process of transforming the specification into a collection of biological building blocks with sufficient information to physically assemble them in the laboratory environment.
- Trumpet generates permutable genetic circuit constructs from the Specification stage designs by interleaving invertase sites at strategic locations within genetic Devices.
- Physical Assembly - Assembling DNA constructs is a series of transformations which take DNA primitives and combine them into Composite Parts. Issues such as efficiency, design cost, and reliability should be taken into account using optimized algorithms and tools.
- Puppetshow assembles these permutable constructs from the Design stage using a Protocol Automation Stack comprising a high-level programming language executable on a liquid handling robot.
- eLabNotebook helps capture data, and schedule resources and lab activities creating a more design automation friendly laboratory environment.
- Test - Finally, the assembled construct must be verified. In the event that it does not work, the design cycle can continue incorporating the knowledge gained at this stage to try to improve the overall process.
- Wet Lab experimentation tested our tools and methodology by beginning to create a framework for investigating transcription factor interactions in Tuberculosis.
Integration
In order to integrate these areas fully together we built upon the Clotho design framework. Clotho is a portable and extensible collection of open-source application software, a concise data model, and a core software development platform for synthetic biological engineering. Being open-source and easily extensible, Clotho is built to address the current and future needs of synthetic biologists. Clotho's data model captures the data associated with synthetic biological parts, devices, systems, and samples, in a unified relational database. Clotho's application software is modularized into "Apps" which all exchange biological data with a database through a uniform interface. Furthermore, Clotho's core platform introduces a concise and simple application programming interface (API) to developers who wish to contribute new applications to Clotho and create customized in-house tools. The API establishes a vocabulary at a level of abstraction familiar to synthetic biologists.
For a more extended overview, check out our talk at the Americas Regional competition:
 "
"
