Team:TzuChiU Formosa/Notebook
From 2011.igem.org
(→Photopaper) |
(→Photopaper) |
||
| (30 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
<div id="wrap"> | <div id="wrap"> | ||
<div id="headerimage"> | <div id="headerimage"> | ||
| - | <img src ="https://static.igem.org/mediawiki/2011/ | + | <img src ="https://static.igem.org/mediawiki/2011/4/47/2011banner.gif" height="150" width="960"> |
</div> | </div> | ||
| Line 30: | Line 30: | ||
=='''Photopaper'''== | =='''Photopaper'''== | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==='''2011.02.24'''=== | ==='''2011.02.24'''=== | ||
| - | '''Discussion:''' | + | '''Discussion:''' |
| - | *Team organization | + | *Team organization[[File:001.jpg|right|500px|caption]] |
*Brain storming | *Brain storming | ||
**paper made by bacteria with add-ons such as colors, fragrance, etc. | **paper made by bacteria with add-ons such as colors, fragrance, etc. | ||
**"light up" the plants for replacing lamp posts. | **"light up" the plants for replacing lamp posts. | ||
| + | |||
==='''2011.03.04'''=== | ==='''2011.03.04'''=== | ||
| Line 47: | Line 45: | ||
**Add-ons: colorful cellulose which produced by bacteria such as beta-carotene | **Add-ons: colorful cellulose which produced by bacteria such as beta-carotene | ||
**information exchange with iGEM 2009 Cambridge team | **information exchange with iGEM 2009 Cambridge team | ||
| + | |||
==='''2011.03.14'''=== | ==='''2011.03.14'''=== | ||
| Line 54: | Line 53: | ||
**Avatar - "light up" the plants by transfecting symbiotic bacteria which cloned into fluorescence gene | **Avatar - "light up" the plants by transfecting symbiotic bacteria which cloned into fluorescence gene | ||
**Eco-friendly warmer - biotic thermal pad | **Eco-friendly warmer - biotic thermal pad | ||
| + | |||
==='''2011.03.23'''=== | ==='''2011.03.23'''=== | ||
| Line 61: | Line 61: | ||
**Option 1 : Culture bacteria which has pigment gene | **Option 1 : Culture bacteria which has pigment gene | ||
**Option 2 : Cellulose-producing bacteria secrete pigment into the medium | **Option 2 : Cellulose-producing bacteria secrete pigment into the medium | ||
| + | |||
==='''2011.03.24'''=== | ==='''2011.03.24'''=== | ||
| Line 68: | Line 69: | ||
** cloning of cellulose gene’s CDS | ** cloning of cellulose gene’s CDS | ||
** the product should operate within E. coli. | ** the product should operate within E. coli. | ||
| + | |||
==='''2011.06.22'''=== | ==='''2011.06.22'''=== | ||
| Line 75: | Line 77: | ||
* -economical and humane way to produce paper in large quantities. | * -economical and humane way to produce paper in large quantities. | ||
* -yeast to be our host | * -yeast to be our host | ||
| + | |||
==='''2011.07.01'''=== | ==='''2011.07.01'''=== | ||
| Line 80: | Line 83: | ||
* Freeze > grin > genome DNA isolation > Cloning = silk protein gene | * Freeze > grin > genome DNA isolation > Cloning = silk protein gene | ||
| + | |||
==='''2011.07.09'''=== | ==='''2011.07.09'''=== | ||
| Line 85: | Line 89: | ||
* the connections between 3 silk proteins : Fibl Fibh P25 | * the connections between 3 silk proteins : Fibl Fibh P25 | ||
* major proteins : H-chain, L-chain, P25 | * major proteins : H-chain, L-chain, P25 | ||
| + | |||
==='''2011.07.15'''=== | ==='''2011.07.15'''=== | ||
| Line 90: | Line 95: | ||
* Due to limited resources and techniques required, the team decided to switch back to the previous project. Paper making ! | * Due to limited resources and techniques required, the team decided to switch back to the previous project. Paper making ! | ||
* However it would be modified to be more innovative and creative. | * However it would be modified to be more innovative and creative. | ||
| + | |||
==='''2011.07.18'''=== | ==='''2011.07.18'''=== | ||
| Line 95: | Line 101: | ||
* Latest project : Photo paper | * Latest project : Photo paper | ||
* cyanobacteria is designed as the host, cellulose made up of the glucose produced by cyanobacteria could be one of the main attraction of the project. | * cyanobacteria is designed as the host, cellulose made up of the glucose produced by cyanobacteria could be one of the main attraction of the project. | ||
| + | |||
==='''2011.07.23'''=== | ==='''2011.07.23'''=== | ||
| Line 102: | Line 109: | ||
* Biobricks from Tokyo 2010 team will be utilized | * Biobricks from Tokyo 2010 team will be utilized | ||
**regulator promoter in order to regulate the secretion of cellulose to solve the aggregation of the bacteria | **regulator promoter in order to regulate the secretion of cellulose to solve the aggregation of the bacteria | ||
| + | |||
==='''2011.09.07'''=== | ==='''2011.09.07'''=== | ||
| Line 117: | Line 125: | ||
______________________________________________________ | ______________________________________________________ | ||
1L→take100ml | 1L→take100ml | ||
| - | |||
+ | + | ||
Yeast Extrat 0.5g | Yeast Extrat 0.5g | ||
| Line 130: | Line 137: | ||
'''Raise E. coli(PSB1C3)''' | '''Raise E. coli(PSB1C3)''' | ||
1.50ml LB+500μl CHLORAMPHENICOL | 1.50ml LB+500μl CHLORAMPHENICOL | ||
| - | 2. 37℃, overnight (14-16hrs) | + | 2.37℃, overnight (14-16hrs) |
</pre> | </pre> | ||
<gallery> | <gallery> | ||
| Line 138: | Line 145: | ||
==='''2011.09.08-13'''=== | ==='''2011.09.08-13'''=== | ||
| - | + | ||
'''Plasmid miniprep kit''' | '''Plasmid miniprep kit''' | ||
| - | PSB1C3 plasmid | + | <pre>PSB1C3 plasmid</pre> |
'''Raise Rhodobacter rubrum''' | '''Raise Rhodobacter rubrum''' | ||
| + | <pre> | ||
1.50ml LB+500μl CHLORAMPHENICOL | 1.50ml LB+500μl CHLORAMPHENICOL | ||
2.37℃, overnight (14-16hrs) | 2.37℃, overnight (14-16hrs) | ||
| Line 157: | Line 165: | ||
'''Raise Gluconacetobacter hansenii''' | '''Raise Gluconacetobacter hansenii''' | ||
1.50ml LB+500μl CHLORAMPHENICOL | 1.50ml LB+500μl CHLORAMPHENICOL | ||
| - | 2. 37℃, overnight (14-16hrs) | + | 2.37℃, overnight (14-16hrs) |
</pre> | </pre> | ||
<gallery> | <gallery> | ||
| Line 169: | Line 177: | ||
'''Digestion check of DNA''' | '''Digestion check of DNA''' | ||
PSB1C3 DNA (control) 3μl | PSB1C3 DNA (control) 3μl | ||
| + | |||
PSB1C3/ EcoR | PSB1C3/ EcoR | ||
| - | DNA | + | DNA 500ng |
10×buffer 5μl | 10×buffer 5μl | ||
BSA 5μl | BSA 5μl | ||
| Line 180: | Line 189: | ||
PSB1C3/ pstⅠ | PSB1C3/ pstⅠ | ||
| - | DNA | + | DNA 500ng |
10×buffer 5μl | 10×buffer 5μl | ||
BSA 5μl | BSA 5μl | ||
| Line 190: | Line 199: | ||
PSB1C3/ EcoRⅠ+pstⅠ | PSB1C3/ EcoRⅠ+pstⅠ | ||
| - | DNA | + | DNA 500ng |
| - | + | 10×buffer 5μl | |
| - | + | BSA 5μl | |
EcoRⅠ 1μl | EcoRⅠ 1μl | ||
pstⅠ 1μl | pstⅠ 1μl | ||
| Line 222: | Line 231: | ||
</gallery> | </gallery> | ||
| - | + | ==='''2011.10.14-02'''=== | |
| - | ==='''2011. | + | |
<pre> | <pre> | ||
'''PCR''' | '''PCR''' | ||
| - | template | + | template DNA 1μl |
5×Buffer 4μl | 5×Buffer 4μl | ||
2.5μM dNTP 1.6μl | 2.5μM dNTP 1.6μl | ||
| Line 236: | Line 244: | ||
______________________________________________________ | ______________________________________________________ | ||
20μl | 20μl | ||
| - | |||
</pre> | </pre> | ||
<gallery> | <gallery> | ||
| Line 260: | Line 267: | ||
<pre> | <pre> | ||
'''Digestion of DNA''' | '''Digestion of DNA''' | ||
| - | acsAB/ | + | acsAB/ XbaⅠ+SpeⅠ |
DNA 10μl | DNA 10μl | ||
10×buffer 5μl | 10×buffer 5μl | ||
BSA 5μl | BSA 5μl | ||
| - | + | XbaⅠ 1μl | |
| - | + | SpeⅠ 1μl | |
ddH2O 28μl | ddH2O 28μl | ||
(+ | (+ | ||
______________________________________________________ | ______________________________________________________ | ||
50μl | 50μl | ||
| - | →37℃ for | + | →37℃ for 16 hr |
'''Digestion of DNA''' | '''Digestion of DNA''' | ||
| - | acsCD/ | + | acsCD/ XbaⅠ+SpeⅠ |
DNA 10μl | DNA 10μl | ||
10×buffer 5μl | 10×buffer 5μl | ||
BSA 5μl | BSA 5μl | ||
| - | + | XbaⅠ 1μl | |
| - | + | SpeⅠ 1μl | |
ddH2O 28μl | ddH2O 28μl | ||
(+ | (+ | ||
______________________________________________________ | ______________________________________________________ | ||
50μl | 50μl | ||
| - | →37℃ for | + | →37℃ for 16 hr |
</pre> | </pre> | ||
| - | |||
==='''2011.09.22'''=== | ==='''2011.09.22'''=== | ||
| Line 291: | Line 297: | ||
'''Ligation of DNA''' | '''Ligation of DNA''' | ||
PSB1C3-acsAB | PSB1C3-acsAB | ||
| - | Vector | + | Vector 3μl |
| - | Insert | + | Insert 14μl |
| - | ligase buffer | + | ligase buffer 2μl |
| - | ligase | + | ligase 1μl |
| - | ddH2O | + | ddH2O -μl |
(+ | (+ | ||
______________________________________________________ | ______________________________________________________ | ||
| - | + | 20μl | |
| - | + | ||
'''Ligation of DNA''' | '''Ligation of DNA''' | ||
| - | + | PSB1A3-acsCD | |
| - | Vector | + | Vector 3μl |
| - | Insert | + | Insert 14μl |
| - | ligase buffer | + | ligase buffer 2μl |
| - | ligase | + | ligase 1μl |
| - | ddH2O | + | ddH2O -μl |
(+ | (+ | ||
______________________________________________________ | ______________________________________________________ | ||
| - | + | 20μl | |
| - | + | ||
</pre> | </pre> | ||
==='''2011.09.23'''=== | ==='''2011.09.23'''=== | ||
| + | <pre> | ||
| + | '''PCR''' | ||
| + | PR0011 promoter 1μl | ||
| + | 5×Buffer 4μl | ||
| + | 2.5μM dNTP 1.6μl | ||
| + | Taq 0.2μl | ||
| + | ddH2O 13.2μl | ||
| + | (+ | ||
| + | ______________________________________________________ | ||
| + | 20μl | ||
| + | </pre> | ||
| + | |||
| + | ==='''2011.09.24'''=== | ||
<pre> | <pre> | ||
'''Transformation of DNA''' | '''Transformation of DNA''' | ||
| Line 332: | Line 349: | ||
Transform into E.coli | Transform into E.coli | ||
LB+Ampicillin+CHLORAMPHENICOL | LB+Ampicillin+CHLORAMPHENICOL | ||
| + | </pre> | ||
| - | |||
| + | ==='''2011.09.24'''=== | ||
| + | <pre> | ||
| + | '''Ligation of DNA''' | ||
| + | PSB1C3-promoter | ||
| + | Vector 3μl | ||
| + | Insert 14μl | ||
| + | ligase buffer 2μl | ||
| + | ligase 1μl | ||
| + | ddH2O -μl | ||
| + | (+ | ||
| + | ______________________________________________________ | ||
| + | 20μl | ||
| + | </pre> | ||
| + | |||
| + | |||
| + | ==='''2011.09.27'''=== | ||
| + | <pre> | ||
| + | '''Digestion of DNA''' | ||
| + | acsCD/ XbaⅠ+SpeⅠ | ||
| + | DNA 10μl | ||
| + | 10×buffer 5μl | ||
| + | BSA 5μl | ||
| + | XbaⅠ 1μl | ||
| + | SpeⅠ 1μl | ||
| + | ddH2O 28μl | ||
| + | (+ | ||
| + | ______________________________________________________ | ||
| + | 50μl | ||
| + | →37℃ for 16 hr | ||
| + | |||
| + | '''Ligation of DNA''' | ||
| + | PSB1C3-acsCD | ||
| + | Vector 3μl | ||
| + | Insert 14μl | ||
| + | ligase buffer 2μl | ||
| + | ligase 1μl | ||
| + | ddH2O -μl | ||
| + | (+ | ||
| + | ______________________________________________________ | ||
| + | 20μl | ||
| + | '''Ligation of DNA''' | ||
| + | PSB1C3-promoter | ||
| + | Vector 3μl | ||
| + | Insert 14μl | ||
| + | ligase buffer 2μl | ||
| + | ligase 1μl | ||
| + | ddH2O -μl | ||
| + | (+ | ||
| + | ______________________________________________________ | ||
| + | 20μl | ||
| + | </pre> | ||
| + | |||
| + | |||
| + | ==='''2011.09.27'''=== | ||
| + | <pre> | ||
| + | '''Ligation of DNA''' | ||
| + | PSB1C3-Cmccax-Ccp 3μl | ||
| + | Insert 14μl | ||
| + | ligase buffer 2μl | ||
| + | ligase 1μl | ||
| + | ddH2O -μl | ||
| + | (+ | ||
| + | ______________________________________________________ | ||
| + | 20μl | ||
| + | |||
| + | </pre> | ||
| + | |||
| + | |||
| + | ==='''2011.09.28'''=== | ||
| + | <pre> | ||
| + | '''Digestion of DNA''' | ||
| + | acsCD/ XbaⅠ+SpeⅠ | ||
| + | DNA 10μl | ||
| + | 10×buffer 5μl | ||
| + | BSA 5μl | ||
| + | XbaⅠ 1μl | ||
| + | SpeⅠ 1μl | ||
| + | ddH2O 28μl | ||
| + | (+ | ||
| + | ______________________________________________________ | ||
| + | 50μl | ||
| + | →37℃ for 16 hr | ||
| + | |||
| + | |||
| + | ==='''2011.09.28'''=== | ||
| + | <pre> | ||
| + | '''Transformation of DNA''' | ||
| + | PSB1C3-Cmccax-Ccp | ||
| + | Transform into E.coli | ||
| + | LB+CHLORAMPHENICOL | ||
| + | </pre> | ||
| + | |||
| + | '''Digestion of DNA''' | ||
| + | <pre> | ||
| + | acsAB/ XbaⅠ+SpeⅠ | ||
| + | DNA 10μl | ||
| + | 10×buffer 5μl | ||
| + | BSA 5μl | ||
| + | XbaⅠ 1μl | ||
| + | SpeⅠ 1μl | ||
| + | ddH2O 28μl | ||
| + | (+ | ||
| + | ______________________________________________________ | ||
| + | 50μl | ||
| + | →37℃ for 16 hr | ||
| + | |||
| + | acsAB/ XbaⅠ+SpeⅠ | ||
| + | DNA 10μl | ||
| + | 10×buffer 5μl | ||
| + | BSA 5μl | ||
| + | XbaⅠ 1μl | ||
| + | SpeⅠ 1μl | ||
| + | ddH2O 28μl | ||
| + | (+ | ||
| + | ______________________________________________________ | ||
| + | 50μl | ||
| + | →37℃ for 16 hr | ||
| + | </pre> | ||
</pre> | </pre> | ||
| Line 350: | Line 485: | ||
File:Outdoors_shooting(3).jpg | File:Outdoors_shooting(3).jpg | ||
</gallery> | </gallery> | ||
| - | |||
| - | |||
==='''2011.06.14'''=== | ==='''2011.06.14'''=== | ||
| Line 444: | Line 577: | ||
File:Heermosi codon meeting(4).jpg | File:Heermosi codon meeting(4).jpg | ||
</gallery> | </gallery> | ||
| + | |||
==='''2011.02.13'''=== | ==='''2011.02.13'''=== | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | '''Every team members propose their ideas for discussion. ''' | |
| - | *Edible bacteria | + | *3 selected project : Edible bacteria, Glowing mushrooms, Combustible bacteria. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
| + | ==='''2011.02.13'''=== | ||
| - | + | *Project selected : Edible bacteria | |
| - | + | ||
| - | * | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | Edible bacteria | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==='''2011.03.21'''=== | ==='''2011.03.21'''=== | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
'''Discussion:''' | '''Discussion:''' | ||
| - | * | + | * sequence of the soybean protein |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| + | ==='''2011.05.06'''=== | ||
| + | * Restriction enzyme cutting site of experiment is not yet confirmed. The transport mechanism of nutrient needs more understanding. | ||
| - | ==='''2011. | + | ==='''2011.05.25'''=== |
| - | + | * Promoter pAMJ399 of lactobacillus’s plasmid is found. | |
| - | + | ==='''2011.06.26'''=== | |
| - | + | * The design of experiment have to discuss with team advisor. Primer have to be customized. | |
| - | * | + | |
| - | + | ||
| - | + | ||
Latest revision as of 17:19, 2 October 2011


Photopaper
2011.02.24
Discussion:
- Team organization
- Brain storming
- paper made by bacteria with add-ons such as colors, fragrance, etc.
- "light up" the plants for replacing lamp posts.
2011.03.04
Discussion:
- Team advisory
- Brain storming
- Add-ons: colorful cellulose which produced by bacteria such as beta-carotene
- information exchange with iGEM 2009 Cambridge team
2011.03.14
Discussion:
- Task Allocation
- Brain storming
- Avatar - "light up" the plants by transfecting symbiotic bacteria which cloned into fluorescence gene
- Eco-friendly warmer - biotic thermal pad
2011.03.23
Discussion:
- Project : paperia
- Option 1 : Culture bacteria which has pigment gene
- Option 2 : Cellulose-producing bacteria secrete pigment into the medium
2011.03.24
Discussion:
- Exp. procedure:
- cloning of cellulose gene’s CDS
- the product should operate within E. coli.
2011.06.22
Discussion:
- Due to some unforseen reason, the team decided to change their project.
- New project: Biojenny
- -economical and humane way to produce paper in large quantities.
- -yeast to be our host
2011.07.01
Discussion:
- Freeze > grin > genome DNA isolation > Cloning = silk protein gene
2011.07.09
Discussion:
- the connections between 3 silk proteins : Fibl Fibh P25
- major proteins : H-chain, L-chain, P25
2011.07.15
Discussion:
- Due to limited resources and techniques required, the team decided to switch back to the previous project. Paper making !
- However it would be modified to be more innovative and creative.
2011.07.18
Discussion:
- Latest project : Photo paper
- cyanobacteria is designed as the host, cellulose made up of the glucose produced by cyanobacteria could be one of the main attraction of the project.
2011.07.23
Discussion:
- system modification to overcome the problems arises during preliminary round
- Biobricks from Tokyo 2010 team will be utilized
- regulator promoter in order to regulate the secretion of cellulose to solve the aggregation of the bacteria
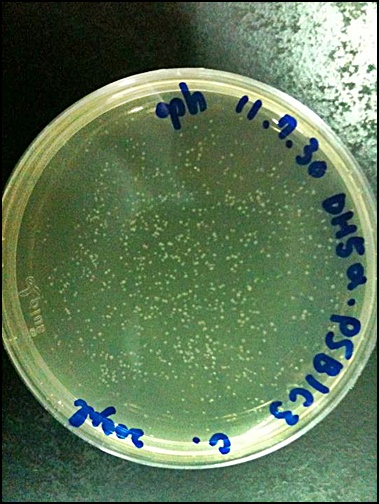
2011.09.07
'''Rhodobacter rudrum medium'''
K2HPO4 1g
NaCl 0.5g
FeSO4.7H2O 0.01g
CaCl2 0.02g
MnCl2.4H2O 0.002g
MgSO4.7H2O 0.2g
NaMO2O4.2H2O 0.01g
ddH2O 998.258 ml
(+
______________________________________________________
1L→take100ml
+
Yeast Extrat 0.5g
Sodium malate
(Sodium succinate dibasic hexohydrate) 5g
NH4Cl 1g
ddH2O 893.5ml
(+
______________________________________________________
1L
'''Raise E. coli(PSB1C3)'''
1.50ml LB+500μl CHLORAMPHENICOL
2.37℃, overnight (14-16hrs)
2011.09.08-13
Plasmid miniprep kit
PSB1C3 plasmid
Raise Rhodobacter rubrum
1.50ml LB+500μl CHLORAMPHENICOL 2.37℃, overnight (14-16hrs)
2011.09.09
'''Raise Gluconacetobacter hansenii''' 1.50ml LB+500μl CHLORAMPHENICOL 2.37℃, overnight (14-16hrs)
2011.09.10-11
'''Digestion check of DNA'''
PSB1C3 DNA (control) 3μl
PSB1C3/ EcoR
DNA 500ng
10×buffer 5μl
BSA 5μl
EcoRⅠ 1μl
ddH2O 29μl
(+
______________________________________________________
50μl
PSB1C3/ pstⅠ
DNA 500ng
10×buffer 5μl
BSA 5μl
pstⅠ 1μl
ddH2O 29μl
(+
______________________________________________________
50μl
PSB1C3/ EcoRⅠ+pstⅠ
DNA 500ng
10×buffer 5μl
BSA 5μl
EcoRⅠ 1μl
pstⅠ 1μl
ddH2O 28μl
(+
______________________________________________________
50μl
→37℃ for 30 mins
'''Digestion of DNA'''
PSB1C3/ EcoRⅠ+pstⅠ
DNA 10μl
10×buffer 5μl
BSA 5μl
EcoRⅠ 1μl
pstⅠ 1μl
ddH2O 28μl
(+
______________________________________________________
50μl
→37℃ for 2 hrs
'''electroelution Purification'''
PSB1C3 backbone
2011.10.14-02
'''PCR'''
template DNA 1μl
5×Buffer 4μl
2.5μM dNTP 1.6μl
10μM F 1μl
10μM R 1μl
Taq 0.2μl
ddH2O 8.8μl
(+
______________________________________________________
20μl
2011.09.15
'''Genome miniprep''' Gluconacetobacter hansenii
2011.09.18
'''Gel/PCR DNA extraction''' Gluconacetobacter hansenii
2011.09.21
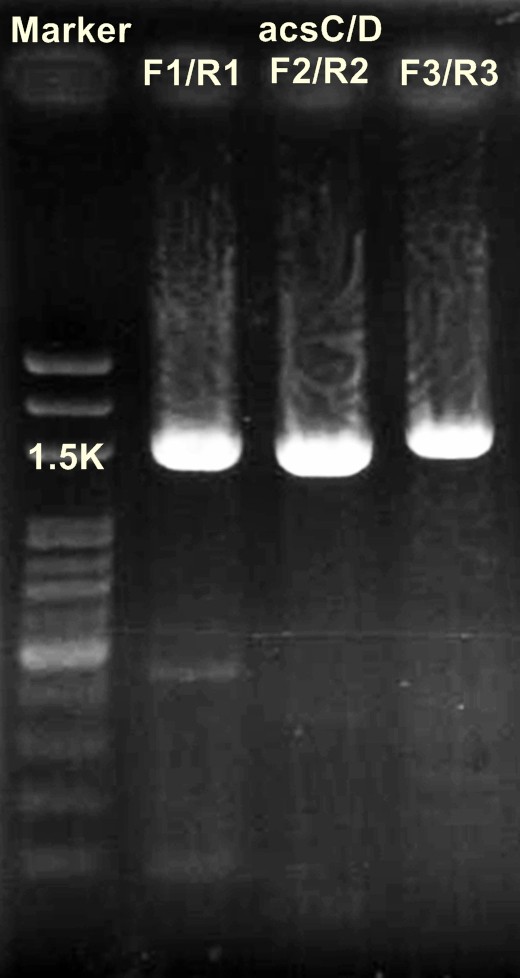
'''Digestion of DNA'''
acsAB/ XbaⅠ+SpeⅠ
DNA 10μl
10×buffer 5μl
BSA 5μl
XbaⅠ 1μl
SpeⅠ 1μl
ddH2O 28μl
(+
______________________________________________________
50μl
→37℃ for 16 hr
'''Digestion of DNA'''
acsCD/ XbaⅠ+SpeⅠ
DNA 10μl
10×buffer 5μl
BSA 5μl
XbaⅠ 1μl
SpeⅠ 1μl
ddH2O 28μl
(+
______________________________________________________
50μl
→37℃ for 16 hr
2011.09.22
'''Ligation of DNA'''
PSB1C3-acsAB
Vector 3μl
Insert 14μl
ligase buffer 2μl
ligase 1μl
ddH2O -μl
(+
______________________________________________________
20μl
'''Ligation of DNA'''
PSB1A3-acsCD
Vector 3μl
Insert 14μl
ligase buffer 2μl
ligase 1μl
ddH2O -μl
(+
______________________________________________________
20μl
2011.09.23
'''PCR'''
PR0011 promoter 1μl
5×Buffer 4μl
2.5μM dNTP 1.6μl
Taq 0.2μl
ddH2O 13.2μl
(+
______________________________________________________
20μl
2011.09.24
'''Transformation of DNA''' PSB1C3-acsAB Transform into E.coli LB+CHLORAMPHENICOL '''Transformation of DNA''' PSB1A3-acsCD Transform into E.coli LB+Ampicillin '''Transformation of DNA''' PSB1C3-acsAB PSB1A3-acsCD Transform into E.coli LB+Ampicillin+CHLORAMPHENICOL
2011.09.24
'''Ligation of DNA'''
PSB1C3-promoter
Vector 3μl
Insert 14μl
ligase buffer 2μl
ligase 1μl
ddH2O -μl
(+
______________________________________________________
20μl
2011.09.27
'''Digestion of DNA'''
acsCD/ XbaⅠ+SpeⅠ
DNA 10μl
10×buffer 5μl
BSA 5μl
XbaⅠ 1μl
SpeⅠ 1μl
ddH2O 28μl
(+
______________________________________________________
50μl
→37℃ for 16 hr
'''Ligation of DNA'''
PSB1C3-acsCD
Vector 3μl
Insert 14μl
ligase buffer 2μl
ligase 1μl
ddH2O -μl
(+
______________________________________________________
20μl
'''Ligation of DNA'''
PSB1C3-promoter
Vector 3μl
Insert 14μl
ligase buffer 2μl
ligase 1μl
ddH2O -μl
(+
______________________________________________________
20μl
2011.09.27
'''Ligation of DNA'''
PSB1C3-Cmccax-Ccp 3μl
Insert 14μl
ligase buffer 2μl
ligase 1μl
ddH2O -μl
(+
______________________________________________________
20μl
2011.09.28
'''Digestion of DNA'''
acsCD/ XbaⅠ+SpeⅠ
DNA 10μl
10×buffer 5μl
BSA 5μl
XbaⅠ 1μl
SpeⅠ 1μl
ddH2O 28μl
(+
______________________________________________________
50μl
→37℃ for 16 hr
==='''2011.09.28'''===
<pre>
'''Transformation of DNA'''
PSB1C3-Cmccax-Ccp
Transform into E.coli
LB+CHLORAMPHENICOL
Digestion of DNA
acsAB/ XbaⅠ+SpeⅠ
DNA 10μl
10×buffer 5μl
BSA 5μl
XbaⅠ 1μl
SpeⅠ 1μl
ddH2O 28μl
(+
______________________________________________________
50μl
→37℃ for 16 hr
acsAB/ XbaⅠ+SpeⅠ
DNA 10μl
10×buffer 5μl
BSA 5μl
XbaⅠ 1μl
SpeⅠ 1μl
ddH2O 28μl
(+
______________________________________________________
50μl
→37℃ for 16 hr
</pre>
CO killer
2011.06.14
Today is our 1st meeting. We are a team of 9 consisting of 2 junior year students and 7 sophomore year students.
Today we discussed about team organization and task allocation. Ting-Ting was given the chance to the team leader while Steven and Li-Yi are the in-charge of system designation. We also plan our schedule so that we could make it in time.
It was quite an awkward meeting because somehow it was like when we were small going to kindergarten the 1st week. Though we study in the same campus, but due to different courses, we barely know each other. But it was very fun making new friends.
2011.06.24
It’s already end of June but we still did not get any professor to be our team advisor. Today Prof. Ingrid from Dept. of Molecular Biology and Human Genetics want to interview us before she promise us to be our team advisor. We present to her our whole project and how our idea comes up. Later on we had a little chat with her on our idea and also the whole iGEM. Finally, she accepts our invitation!
Team organization - checked !
2011.06.27
Summer is finally here! Weather is so hot and humid.
We had lunch together before we start our meeting. We were solving the problem of carbon dioxide produced by oxidation of carbon monoxide. The most ideal method is to use either plants or algae to convert the CO2 to biomass which is then processed to produce biofuels. However after long discussion with Professor Chen, we realized that we couldn’t reach that goal in such short period but we can design the host bacteria to use the Carbon Monoxide as one of the carbon source. It’s not a bad idea after all =D
2011.06.30
We all did some research on how Carbon Monoxide affects human body and what concentration level of CO is a concern. Carbon monoxide exposure levels are measured in parts per million or ppm. Steven and Joanne found out that different country has different standards. It was quite confusing because the figure is of big difference. However, after discussion with professor we knew that the risk allowance is also included in it, which is why the safety limits are not the same. Therefore, we can know conclude that, the safety standard is affected by the ppm of carbon monoxide and also the risk allowance.
We also start to do our powerpoint presentation slide which we going to show during the preliminary round.
Everyone is so focus now! Because we believed together we can achieve more. Hopefully we can keep the spirit up =D
2011.07.04
Today we had our meeting in the afternoon. The weather is so hot. It was like barbeque ourselves under the sun so we decided not to have meeting in the afternoon anymore. The topic today is the oxidation of CO to CO2 – we found out that Rhodospirillum rubrum can undergo Calvin Cycle to obtain their energy from sunlight and use CO2 as their carbon source, however cyanobacteria or other bacteria is needed for CO2 fixation and production of biofuels. But it would be much more sophiscated if we add on the cyanobacteria into the system. Hmmm … looks like we have to improve more on our system design !
2011.07.05
We talked to the microbiologist Prof. Chen CY yesterday evening and stunned us with 3 questions. The time factor, the mutation factor and the gene expression rate. The experiment of CO2 fixation requires at least half a year to complete so we probably couldn’t just simply add in our project. Besides that, we also have to think of the mutation of the gene. If the gene mutated then the product would be useless. And the most important is the rate of the protein to turn fluorescent. Lastly, if the gene size is getting bigger it would be more difficult to insert into the plasmid. Consequently, we would need more time in cloning and that would be time-costing. Uhh … we really need to find the resolution soon ! Any way, Prof. Ingrid is really very busy. We have not see her for the past 2 weeks. Gotta check out with her lab assistant.
2011.07.06
Today we had meeting in the afternoon again. Thanks god the weather is windy. We downloaded all the sequences and CDS of the gene we need. On the other hand we also search the sources of R. Rubrum which we planned to use in our project. There is a few proffesor in taiwan doing the research about R.Rubrum. So Joanne will help the letter to them to request a sample of it. And the worst case we can buy it from biotech corporation. Steven need somebody to be the in-charge of wikipedia. Leo volunteered. Found out that 2 enzymes are available : CODH–Nickel, OM5 –Molybdenum; CODH is has a higher rate of binding with CO, but OM5 can bind with CO in enormous amount at one time. Say no to procrastination ! Next meeting on Friday. A day off tomorrow =D
2011.07.09
Discussion regarding the presentation slide during the preliminary round. Task allocated to all the members. While Joanne and Steven had to write the project proposal ASAP. Not much were done today. But TGIF !! Had some photo session after the meeting. Joanne go crazy with the effect thingy on the MacBook. Stressed up after one week of intensive meeting.
2011.07.12
We are all so excited today because we having a photoshooting session this afternoon. We asked another classmate of us, Jason to be our professional photographer. It looks like all of us are good model. It started to drizzle when we were moving to another spot. Too bad we only can do indoor photoshoot. Nevermind, we can have another one in the laboratory next time. In the evening, we also had a meeting with Prof. Ingrid. We did a rehearsal about our preliminary round with her. We really have to speed up to improve our slideshow so that we could stand a chance in the coming competition.
2011.07.15
We had our second rehearsal. And we checked the email, too bad there’s no reply from the 3 professors who we send them the request letter. I think we should stalk the professor in the facebook.
2011.07.18
We had a quick briefing with Steven today. We just got the instructions to attend a meeting with our organizer on 26th of July.
Our preliminary is fixed on 22th July 12 noon. It’s only a week left, we have to work hard !!
2011.07.20
It’s the final rehearsal before our competition. Everyone is well-prepared already. Today we even wore our formal wear to the rehearsal. Steven and Leo look so smart. Practice makes perfect, now everyone can explain the slide very well. I believed that together we can achieve more.
2011.07.25
We make it to the next round! It was the big day today. The professors are impressed by our idea and really like ours creativity. However, our main problem is still the system design which is that the concentration of Carbon Monoxide which will be the inducer of the operon. Also, another problem is that we have to renew the host bacteria regularly.
2011.07.27
Finally, the deputy director of the national museum of marine biology and aquarium replied us. Too bad he doesn’t have the sample of R.Rubrum. Anyway, we’re quite motivated by his words. Thanks Prof. CS Chen
Helmersi Codon
2011.02.13
Every team members propose their ideas for discussion.
- 3 selected project : Edible bacteria, Glowing mushrooms, Combustible bacteria.
2011.02.13
- Project selected : Edible bacteria
2011.03.21
Discussion:
- sequence of the soybean protein
2011.05.06
- Restriction enzyme cutting site of experiment is not yet confirmed. The transport mechanism of nutrient needs more understanding.
2011.05.25
- Promoter pAMJ399 of lactobacillus’s plasmid is found.
2011.06.26
- The design of experiment have to discuss with team advisor. Primer have to be customized.
 "
"