Team:Potsdam Bioware/Project/Details Microviridin
From 2011.igem.org
(→Modularization of the mdn gene cluster) |
(→Modularization of the mdn gene cluster) |
||
| (49 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
==Microviridin== | ==Microviridin== | ||
=== Introduction === | === Introduction === | ||
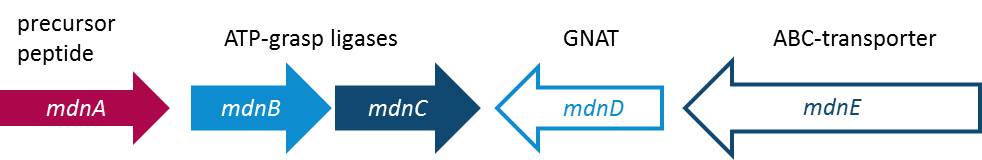
| - | Cyanobacteria are known as the blue-green-algae because their living space is water and their feeding mechanism is photosynthetic. Some of | + | Cyanobacteria are known as the blue-green-algae because their living space is water and their feeding mechanism is photosynthetic. Cyanobacteria are know for special and unique metabolites. Some of these metabolites are part of the microviridin family. Special attributes of microviridins are their occurrence as tricyclic depsipeptides, which means peptides with one or more amide-bonds replaced by ester-bonds, unparalleled cage-like architecture, and the potential to inhibit proteases. Microviridin B, for example, has been reported to function as an elastase inhibitor what could be used as an therapeutic attempt to fix the out-of-control function of elastase in lung emphysema. In such a case, it is beyond debate how important it is to figure out the biosynthesis of this peptide. The research group of Prof. Dittmann reported the composition of the gene cluster from ''Micocystis aeruginosa'' NIES298 expressing Microviridin B. Moreover they have been convinced that there is an unique biosynthetic mechanism for microviridins in <i>Microcystis</i> strains. Unusual for depsipeptides, they discovered that Microviridins are ribosomally synthesized. The ''mdn'' gene cluster contains the gene ''mdnA'', encoding for the putative precursor peptide of the microvirdin, two genes encoding ATP-grasp-type ligases ''mdnB'' and ''mdnC'', an ABC transporter encoding gene ''mdnE'' as well as one encoding an N-acetyltransferase of the GNAT family ''mdnD'' (Ziemert et al., 2008). In a recent study, they report the discovery of an existing natural diversity of microviridin precursors genes in set of closely related <i>Mycrocystis</i> laboratory strains. To give evidence of their discovery, they identified and characterized a new Microviridin L from the strain ''Mycrocystis aeruginosa'' NIES843 (Ziemert et al., 2010). |
| - | All in all the microviridins | + | All in all the microviridins are build up through a gene cluster of five mdn-components each fulfilling one specific function. It was shown that the peptide is only correctly processed in ''E. coli'' if the entire cluster ''mdnABCDE'' is expressed. In addition, the new discovery is indicating some flexibility of the microviridin ligases with an open window for the size of the natural microviridins, and thus, microviridin function and for potential therapeutic benefit that needs to be discovered. The following illustration is going to give you a better overview (Fig. 1). |
| - | [[File:UP_MV_introduction_mdn.jpg|center| | + | [[File:UP_MV_introduction_mdn.jpg|center|525px|thumb|'''Figure 1:''' overview of the mdn gene cluster (GNAT: GCN5-related N-acetyltransferase)]] |
<br><br> | <br><br> | ||
=== Modularization of the ''mdn'' gene cluster=== | === Modularization of the ''mdn'' gene cluster=== | ||
| - | Using two vectors (pARW071 and pARW089, respectively) containing the ''mdn'' biosynthetic gene cluster, we designed PCR primers for the amplification of the ''mdn genes''. Using these primers, we obtained the gene fragments of ''mdnA'', ''mdnB'', ''mdnC'', ''mdnD'', ''mdnE'' and for the whole cluster, respectively. Due to the sophisticated incorporation of the sequences of the iGEM restriction enzymes into the primer sequence our BioBricks comply with the BioBrick standards. | + | Using two vectors (pARW071 and pARW089, respectively) containing the ''mdn'' biosynthetic gene cluster, we designed PCR primers for the amplification of the ''mdn genes''. Using these primers, we obtained the gene fragments of ''mdnA'', ''mdnB'', ''mdnC'', ''mdnD'', ''mdnE'' and for the whole cluster, respectively. Due to the sophisticated incorporation of the recognition sequences of the iGEM restriction enzymes into the primer sequence our BioBricks comply with the BioBrick standards. |
The following image (Fig. 2) shows the generic construction of the BioBrick carrying the'' mdnA gene''. | The following image (Fig. 2) shows the generic construction of the BioBrick carrying the'' mdnA gene''. | ||
| Line 15: | Line 15: | ||
[[File:UP_modularization_mdnA_cloning.png|center|500px|thumb|'''Figure 2:''' Cloning Scheme of mdnA-BioBrick (''Cm'' - gene for chloramphenicol resistance)]] | [[File:UP_modularization_mdnA_cloning.png|center|500px|thumb|'''Figure 2:''' Cloning Scheme of mdnA-BioBrick (''Cm'' - gene for chloramphenicol resistance)]] | ||
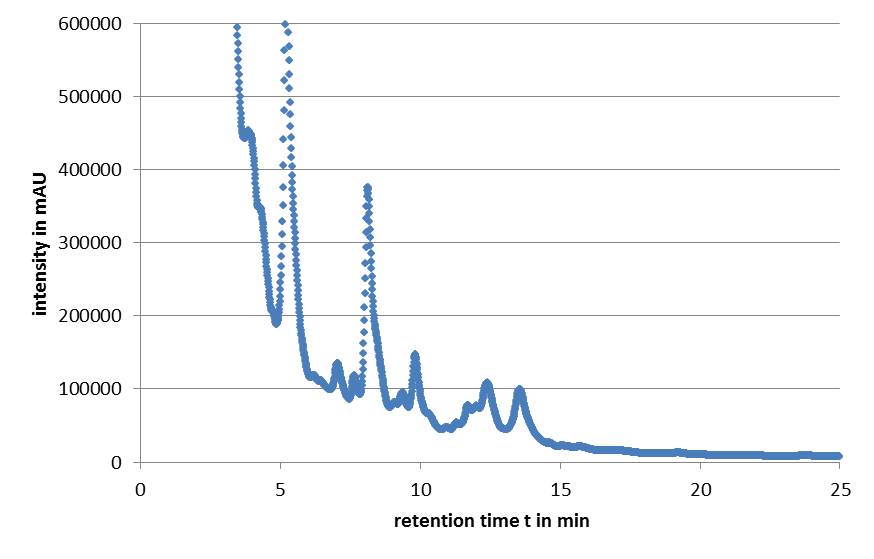
| - | Microviridin production in ''E. coli'' cells expressing the'' mdn genes'' was monitored by | + | Microviridin production in ''E. coli'' cells expressing the'' mdn genes'' was monitored by reversed phase HPLC. Qualitative analysis involved running of a standard that contained the target analytes. The vectors pARW071 and pARW089 served as controls in our experiments, because these vectors contain the original mdn-cluster. We could use the retention time as a way to determine the presence of the microviridin production in other samples. HPLC analysis of the purified compound yielded a high peak with a retention time of approximately 5 min. Minor peaks could be detected during the following 7 min (Fig. 3). |
| - | [[File:UP_modularization_HPLC.png|center|500px|thumb|'''Figure 3:''' HPLC chromatogram of | + | [[File:UP_modularization_HPLC.png|center|500px|thumb|'''Figure 3:''' HPLC chromatogram of MdnA]] |
All HPLC chromatograms of the isolated mdnA showed reliable peaks with a retention time of 5 min together with a number of following minor peaks. | All HPLC chromatograms of the isolated mdnA showed reliable peaks with a retention time of 5 min together with a number of following minor peaks. | ||
| - | Fractions of the peaks were sampled and the identity of microviridin and also the presence of cyclization, which is important for the activity, was | + | Fractions of the peaks were sampled and the identity of microviridin and also the presence of cyclization, which is important for the activity, was confirmed with mass spectrometry (Fig. 4). By using a mass spectrometer it is possible to determine both the elemental composition of a sample and deduce the chemical structure of molecules. |
| - | [[File: | + | [[File:UP_modularization_MSII.gif|center|300px|thumb|'''Figure 4:''' Mass Spectrometry Data]] |
| + | <br><br> | ||
| - | === Generating | + | === Generating ''mdnA'' gene libraries === |
In searching for optimized and novel microviridin peptides, libraries of mutated microviridins were established. Therefore a number of sites in the amino acid sequence of the precursor peptide (MdnA) were chosen. | In searching for optimized and novel microviridin peptides, libraries of mutated microviridins were established. Therefore a number of sites in the amino acid sequence of the precursor peptide (MdnA) were chosen. | ||
| - | In Microcystis, microviridins are synthesized from a ribosomal precursor peptide (MdnA). Therefore the microviridin gene cluster is necessary. This 6.5 kb biosynthesis gene cluster includes two genes (mdnB and mdnC), which encodes ATP-grasp-type ligases and further the mdnE gene encoding an ABC transporter as well as mdnD gene encoding a N-acetyltransferase (Ziemert et al., 2008 | + | In <i>Microcystis</i>, microviridins are synthesized from a ribosomal precursor peptide (MdnA). Therefore the microviridin gene cluster is necessary. This 6.5 kb biosynthesis gene cluster includes two genes (''mdnB'' and ''mdnC''), which encodes ATP-grasp-type ligases and further the ''mdnE'' gene encoding an ABC transporter as well as'' mdnD'' gene encoding a N-acetyltransferase (Ziemert et al., 2008). In consequence the biosynthesis gene cluster genes ''mdnB'', ''mdnC'', ''mdnD'' and ''mdnE'' cannot be mutated. Otherwise only misprocessed and non-function variants of microviridin occur (Ziemert et al., 2008). Thus merely the modification of the ''mdnA'' gene can lead to effective changes in the microviridin structure and function. <br> |
| - | [[File: | + | [[File:UP_lib2_fig.png|center|500px|thumb|'''Figure 5:''' Molecular organization of the ribsosomal precursor peptide (MdnA) of microviridin. MdnA consists of a leader and a core peptide. The N-terminal leader peptide contains highly conserved double glycine motifs. Whereas in the C-terminal 14 amino acid sized core peptide sequence shows variations. Modified from Ziemert et al. (2008) and Ziemert et al. (2010).]] |
| - | The precursor peptide MdnA consists of a leader peptide and a core peptide (Fig. 5). Ziemert et al. (2008) has shown that the N-terminal leader peptide contains highly conserved double glycine motifs (Ziemert et al. 2008 | + | The precursor peptide MdnA consists of a leader peptide and a core peptide (Fig. 5). Ziemert et al. (2008) has shown that the N-terminal leader peptide contains highly conserved double glycine motifs (Ziemert et al. 2008). However remarkable variations were found in region, which encodes the 14-amino acid sequence of the C-terminal microviridin core peptide (Ziemert et al. 2010). Consequently, mutations in the MdnA core peptide seem to be the most promising option for modification of microviridin. For cyclization, the amino acid side chains of the core peptide form omega-ester and omega-amino bonds. One loop is formed between threonine and aspartic acid, the second loop between lysine and aspartic acid and the third loop between serine and glutamic acid (Fig. 5). The sequence of this loop forming amino acids constitute an exception while mutation of the ''mdnA'' gene (Ziemert et al., 2008 and 2010). |
| - | To find optimized and novel microviridin variants, several libraries carrying modifications in the sequence of the MdnA core peptide were established. These libraries were generated by randomized oligonucleotide synthesis. Therefore a forward oligonucleotide displaying a part of conserved leader peptide sequence was built. Further a reverse oligonucleotide was created. The first part of this oligonucleotide is 20 bases, which overlap with the forward oligonucleotide and which is necessary for hybridization of both, forward and reverse, oligonucleotides. In the middle modified bases | + | To find optimized and novel microviridin variants, several libraries carrying modifications in the sequence of the MdnA core peptide were established. These libraries were generated by randomized oligonucleotide synthesis. Therefore a forward oligonucleotide displaying a part of conserved leader peptide sequence was built. Further a reverse oligonucleotide was created. The first part of this oligonucleotide is 20 bases, which overlap with the forward oligonucleotide and which is necessary for hybridization of both, forward and reverse, oligonucleotides. In the middle of the sequence modified bases were inserted. For cloning, the forward oligonucleotide starts with the blunt restricted site of <i>Sfo</i>I. This results therein that the oligonucleotide can ligate in the <i>Sfo</i>I restricted vector without previous digest of the oligonucleotide. The last part of the reverse oligonucleotides displays an <i>Aat</i>II restriction site for cloning. Digestion with <i>Aat</i>II results in sticky ends, thus wrong way round insertion of the oligonucleotide is impossible. Forward and reverse oligonucleotides were combined by performing a fill-in reaction. |
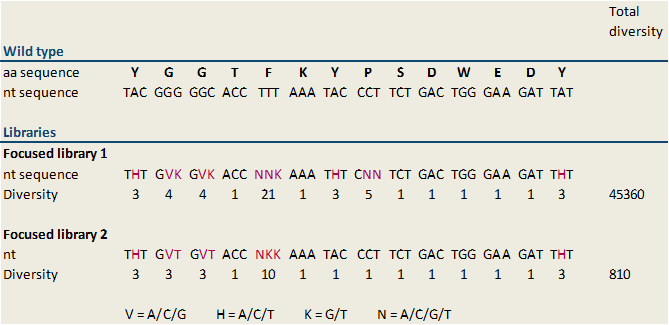
Because we designed several libraries with different diversity rates, the modified bases, which are inserted in the middle of the reverse primer, vary. All of our generated libraries contain mutations of the sequence of the MdnA core peptide at several sites, but not at the loop forming sites. Thus we are thinking that cyclization of microviridin happens accurately. A library with a diversity of 45,360 (focused library 1) and a second focused library showing a minor diversity of 810 (focused library 2) were designed. The included sequence modifications plus the diversity of our libraries are shown in figure 6. In our further studies we worked on focused library 2 primarily. In this library the nucleotide sequences encoding glycine residues were changed in GVK in all cases (Figure 6). GVK stands for guanine at the first position, adenine, cytosine or guanine at the second position and further thymine at the third position of the codon. These three possible codons encode for the amino acids alanine, aspartic acid and glycine. The nucleotide sequence N-terminal tyrosine residue was changed in THT describing thymine (first position), adenine, cytosine or thymine at the second position and thymine (third position). These codons encode isoleucine, leucine and phenylalanine. Additionally, the sequence of phenylalanine in the core peptide sequence was shift to NNK. NKK stands for one of the four nucleotides at the first position, and guanine or thymine at the second and third position. Consequently, this codon encodes the amino acids arginine, glycine, tryptophan, methionine, leucine, valine, serine, cysteine, isoleucine and phenylalanine. | Because we designed several libraries with different diversity rates, the modified bases, which are inserted in the middle of the reverse primer, vary. All of our generated libraries contain mutations of the sequence of the MdnA core peptide at several sites, but not at the loop forming sites. Thus we are thinking that cyclization of microviridin happens accurately. A library with a diversity of 45,360 (focused library 1) and a second focused library showing a minor diversity of 810 (focused library 2) were designed. The included sequence modifications plus the diversity of our libraries are shown in figure 6. In our further studies we worked on focused library 2 primarily. In this library the nucleotide sequences encoding glycine residues were changed in GVK in all cases (Figure 6). GVK stands for guanine at the first position, adenine, cytosine or guanine at the second position and further thymine at the third position of the codon. These three possible codons encode for the amino acids alanine, aspartic acid and glycine. The nucleotide sequence N-terminal tyrosine residue was changed in THT describing thymine (first position), adenine, cytosine or thymine at the second position and thymine (third position). These codons encode isoleucine, leucine and phenylalanine. Additionally, the sequence of phenylalanine in the core peptide sequence was shift to NNK. NKK stands for one of the four nucleotides at the first position, and guanine or thymine at the second and third position. Consequently, this codon encodes the amino acids arginine, glycine, tryptophan, methionine, leucine, valine, serine, cysteine, isoleucine and phenylalanine. | ||
[[File:UP_lib2_fig2.png|center|500px|thumb|'''Figure 6:''' Design of libraries based on the nucleotide sequence of the ribosomal precursor peptide (MdnA). To different libraries, focused library 1 and focused library 2 were generated by randomised oligonucleotide synthesis. The modifications compared to the wild type sequence are highlighted. The definition of the used code is also shown. The first libray (focused library 1) displays a diversity of 45360 and the second, focused library 2 a diversity of 810. In the further studies focused library 2 was used to find optimized microviridin variants. ]] | [[File:UP_lib2_fig2.png|center|500px|thumb|'''Figure 6:''' Design of libraries based on the nucleotide sequence of the ribosomal precursor peptide (MdnA). To different libraries, focused library 1 and focused library 2 were generated by randomised oligonucleotide synthesis. The modifications compared to the wild type sequence are highlighted. The definition of the used code is also shown. The first libray (focused library 1) displays a diversity of 45360 and the second, focused library 2 a diversity of 810. In the further studies focused library 2 was used to find optimized microviridin variants. ]] | ||
| - | To construct this library a fill-in reaction using the designed forward and reverse oligonucleotides was performed. Subsequently resulting randomized oligonucleotide was digested with the restriction enzyme | + | To construct this library a fill-in reaction using the designed forward and reverse oligonucleotides was performed. Subsequently resulting randomized oligonucleotide was digested with the restriction enzyme <i>Aat</i>II. In addition, the vector pUP089 was digested with the restriction enzymes <i>Aat</i>II and <i>Sfo</i>I. After ligation of fragments, randomized oligonucleotide and vector, the focused library 2 (ligation product) was transformed in chemocompetent ''E. coli'' XL1-Blue cells. This procedure was done several times and a library size of 1233 colonies was reached. |
[[File:UP_lib2_fig3.png|center|500px|thumb|'''Figure 7:''' Confirmation of focused library 2. Different clones of the generated focused library 2 were sequenced. Alignments with the wild type sequence (reference sequence) were performed. As shown (for a selection of clones) in all of our sequenced samples the requested modifications were established.]] | [[File:UP_lib2_fig3.png|center|500px|thumb|'''Figure 7:''' Confirmation of focused library 2. Different clones of the generated focused library 2 were sequenced. Alignments with the wild type sequence (reference sequence) were performed. As shown (for a selection of clones) in all of our sequenced samples the requested modifications were established.]] | ||
For confirmation of the focused library 2, sequencing analysis of a number of clones was performed. In all of our sequenced samples the requested modifications were established (figure 7). A sequence logo was generated for the 14 amino acid sized core peptide of MdnA. This sequence logo, which is illustrated in figure 8, indicates that the desired sites stay conserved. The others are modified. | For confirmation of the focused library 2, sequencing analysis of a number of clones was performed. In all of our sequenced samples the requested modifications were established (figure 7). A sequence logo was generated for the 14 amino acid sized core peptide of MdnA. This sequence logo, which is illustrated in figure 8, indicates that the desired sites stay conserved. The others are modified. | ||
[[File:UP_lib2_fig4.png|center|500px|thumb|'''Figure 8:''' Sequence logo plot of focused library 2. The 14 amino acid sized core peptide of MdnA was sequenced. The sequence of several clones was used to generate a sequence logo. This indicates that the desired sites stay conserved. The others nucleotides and resulting amino acids are modified. ]] | [[File:UP_lib2_fig4.png|center|500px|thumb|'''Figure 8:''' Sequence logo plot of focused library 2. The 14 amino acid sized core peptide of MdnA was sequenced. The sequence of several clones was used to generate a sequence logo. This indicates that the desired sites stay conserved. The others nucleotides and resulting amino acids are modified. ]] | ||
| - | The generated and modified focused library 2 should be used as basis for in | + | The generated and modified focused library 2 should be used as basis for <i>in vivo</i> selection and phage display screening. Furthermore we purpose the objective to search for optimized and novel microviridins by using this library. |
| + | <br><br> | ||
=== Expression Backbones === | === Expression Backbones === | ||
In a subtask of our work we wanted to construct auxiliary expression backbones with inducible promotors. In contrast to constitutive systems inducible systems only express the protein of interest after adding an inducer, which can be for example AHL or arabinose. | In a subtask of our work we wanted to construct auxiliary expression backbones with inducible promotors. In contrast to constitutive systems inducible systems only express the protein of interest after adding an inducer, which can be for example AHL or arabinose. | ||
| - | We decided to construct IPTG- and arabinose-inducible systems. Therefore we amplified the promotor region and fused it to a reporter gene, which is constituted by YFP. Using this we have the option to verify the presence of the inducible promotor and also the function of the induction process by fluorescence. Our idea was to use the reporter gene as a placeholder. Via restriction enzyme digestion using the iGEM restriction enzyme | + | We decided to construct IPTG- and arabinose-inducible systems. Therefore we amplified the promotor region and fused it to a reporter gene, which is constituted by YFP. Using this we have the option to verify the presence of the inducible promotor and also the function of the induction process by fluorescence. Our idea was to use the reporter gene as a placeholder. Via restriction enzyme digestion using the iGEM restriction enzyme sites you will be able to replace the reporter gene with your gene of interest. As vector backbone we made use of the pSB1A3, which has ampicillin resistance (Fig. 9). The constructs using pSB1K3 (kanamycin resistance) and pSB1C3 (chloramphenicol resistance) are still on the anvil. |
| - | [[File:UP_expression_backbones_cloning.png|center|500px|thumb|'''Figure | + | [[File:UP_expression_backbones_cloning.png|center|500px|thumb|'''Figure 9:''' Cloning Scheme of Expression Backbones. ''amp'' - gene for ampicillin resistance]] |
| - | We successfully tested the IPTG- and arabinose-inducible system (Fig | + | We successfully tested the IPTG- and arabinose-inducible system (Fig 10). Using fluorescence microscopy we were able to detect the YFP-expression after an induction time of approximately 1.5 hrs. To prove the hypothesis we opposed an induced sample with a non-induced control for each promotor. By use of brightfield combined with differential interference contrast microscopy we traced the ''E. coli'' cells and switched then to the YFP detecting channel to investigate the fluorescence of these cells. |
| - | [[File:UP_Expression_Backbones_Fluorescence.png|center|800px|thumb|'''Figure | + | [[File:UP_Expression_Backbones_Fluorescence.png|center|800px|thumb|'''Figure 10:''' Fluorescence Microscopy. control - not induced; induced - induction with IPTG or Arabinose, respectively]] |
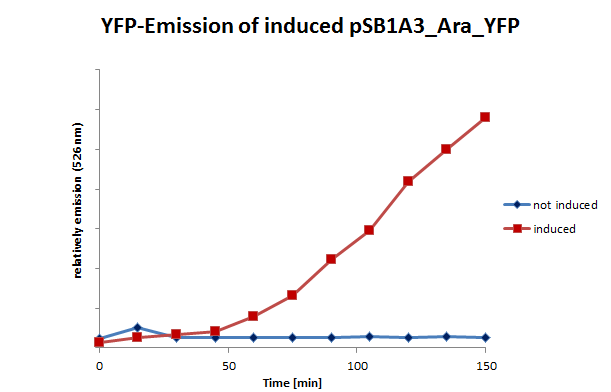
| - | We also tested the inducible systems by using fluorescence spectroscopy. For this experiment we induced both systems at a time when the cultures were located in phase of exponential growth. After inducing, the cultures were analyzed by fluorescence spectroscopy. In this analyze the cells were | + | We also tested the inducible systems by using fluorescence spectroscopy. For this experiment we induced both systems at a time when the cultures were located in phase of exponential growth. After inducing, the cultures were analyzed by fluorescence spectroscopy. In this analyze the cells were excited by 500 nm. The resultant emission was measured in a spectrum between 510 and 580 nm. (Fig 11;12) |
| + | <table><td>[[File:UP_psB1A3_Ara_YFPII.png|left|410px|thumb|'''Figure 11:''' Emission (526nm) of pSB1A3_Ara_YFP and control – '' E.coli'' cells (including pSB1A3_Ara_YFP) were induced and measured at certain moments.]] </td> <td>[[File:UP_psB1A3_IPTG_YFPII.png|right|410px|thumb|'''Figure 12:''' Emission (526nm) of induced pSB1A3_lac_YFP and control – ''E.coli'' cells (including pSB1A3_lac_YFP) were induced and measured at certain moments.]]</td> | ||
| + | </table>In contrast to the control of the arabinose inducible system the Lac-control also shows fluorescence. This demonstrates the leaky <i>Lac</i>-promotor, what is also a scientifically proven fact. A leaky promotor means that the promotor is not regulated in a leakproof manner. Even in repressed conditions the transcript can arise. | ||
| + | The lacking of the <i>LacI</i> gene on the vector is the reason for expression of YFP in both controls. For further research in this field it is necessary to clone the <i>LacI</i> gene inside the constructed vector. | ||
| + | <br><br> | ||
| - | + | ===References=== | |
| + | Ziemert, N., Ishida, K., Liaimer, A., Hertweck, C. & Dittmann, E. (2008). Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angewandte Chemie (International ed. in English) 47, 7756-9 | ||
| - | + | Ziemert, N., Ishida, K., Weiz, A., Hertweck, C. & Dittmann, E. (2010). Exploiting the natural diversity of microviridin gene clusters for discovery of novel tricyclic depsipeptides. Applied and environmental microbiology 76, 3568-74 | |
| - | + | <br><br> | |
| - | + | ||
Latest revision as of 03:27, 29 October 2011
Contents |
Microviridin
Introduction
Cyanobacteria are known as the blue-green-algae because their living space is water and their feeding mechanism is photosynthetic. Cyanobacteria are know for special and unique metabolites. Some of these metabolites are part of the microviridin family. Special attributes of microviridins are their occurrence as tricyclic depsipeptides, which means peptides with one or more amide-bonds replaced by ester-bonds, unparalleled cage-like architecture, and the potential to inhibit proteases. Microviridin B, for example, has been reported to function as an elastase inhibitor what could be used as an therapeutic attempt to fix the out-of-control function of elastase in lung emphysema. In such a case, it is beyond debate how important it is to figure out the biosynthesis of this peptide. The research group of Prof. Dittmann reported the composition of the gene cluster from Micocystis aeruginosa NIES298 expressing Microviridin B. Moreover they have been convinced that there is an unique biosynthetic mechanism for microviridins in Microcystis strains. Unusual for depsipeptides, they discovered that Microviridins are ribosomally synthesized. The mdn gene cluster contains the gene mdnA, encoding for the putative precursor peptide of the microvirdin, two genes encoding ATP-grasp-type ligases mdnB and mdnC, an ABC transporter encoding gene mdnE as well as one encoding an N-acetyltransferase of the GNAT family mdnD (Ziemert et al., 2008). In a recent study, they report the discovery of an existing natural diversity of microviridin precursors genes in set of closely related Mycrocystis laboratory strains. To give evidence of their discovery, they identified and characterized a new Microviridin L from the strain Mycrocystis aeruginosa NIES843 (Ziemert et al., 2010). All in all the microviridins are build up through a gene cluster of five mdn-components each fulfilling one specific function. It was shown that the peptide is only correctly processed in E. coli if the entire cluster mdnABCDE is expressed. In addition, the new discovery is indicating some flexibility of the microviridin ligases with an open window for the size of the natural microviridins, and thus, microviridin function and for potential therapeutic benefit that needs to be discovered. The following illustration is going to give you a better overview (Fig. 1).
Modularization of the mdn gene cluster
Using two vectors (pARW071 and pARW089, respectively) containing the mdn biosynthetic gene cluster, we designed PCR primers for the amplification of the mdn genes. Using these primers, we obtained the gene fragments of mdnA, mdnB, mdnC, mdnD, mdnE and for the whole cluster, respectively. Due to the sophisticated incorporation of the recognition sequences of the iGEM restriction enzymes into the primer sequence our BioBricks comply with the BioBrick standards.
The following image (Fig. 2) shows the generic construction of the BioBrick carrying the mdnA gene.
Microviridin production in E. coli cells expressing the mdn genes was monitored by reversed phase HPLC. Qualitative analysis involved running of a standard that contained the target analytes. The vectors pARW071 and pARW089 served as controls in our experiments, because these vectors contain the original mdn-cluster. We could use the retention time as a way to determine the presence of the microviridin production in other samples. HPLC analysis of the purified compound yielded a high peak with a retention time of approximately 5 min. Minor peaks could be detected during the following 7 min (Fig. 3).
All HPLC chromatograms of the isolated mdnA showed reliable peaks with a retention time of 5 min together with a number of following minor peaks.
Fractions of the peaks were sampled and the identity of microviridin and also the presence of cyclization, which is important for the activity, was confirmed with mass spectrometry (Fig. 4). By using a mass spectrometer it is possible to determine both the elemental composition of a sample and deduce the chemical structure of molecules.
Generating mdnA gene libraries
In searching for optimized and novel microviridin peptides, libraries of mutated microviridins were established. Therefore a number of sites in the amino acid sequence of the precursor peptide (MdnA) were chosen.
In Microcystis, microviridins are synthesized from a ribosomal precursor peptide (MdnA). Therefore the microviridin gene cluster is necessary. This 6.5 kb biosynthesis gene cluster includes two genes (mdnB and mdnC), which encodes ATP-grasp-type ligases and further the mdnE gene encoding an ABC transporter as well as mdnD gene encoding a N-acetyltransferase (Ziemert et al., 2008). In consequence the biosynthesis gene cluster genes mdnB, mdnC, mdnD and mdnE cannot be mutated. Otherwise only misprocessed and non-function variants of microviridin occur (Ziemert et al., 2008). Thus merely the modification of the mdnA gene can lead to effective changes in the microviridin structure and function.

The precursor peptide MdnA consists of a leader peptide and a core peptide (Fig. 5). Ziemert et al. (2008) has shown that the N-terminal leader peptide contains highly conserved double glycine motifs (Ziemert et al. 2008). However remarkable variations were found in region, which encodes the 14-amino acid sequence of the C-terminal microviridin core peptide (Ziemert et al. 2010). Consequently, mutations in the MdnA core peptide seem to be the most promising option for modification of microviridin. For cyclization, the amino acid side chains of the core peptide form omega-ester and omega-amino bonds. One loop is formed between threonine and aspartic acid, the second loop between lysine and aspartic acid and the third loop between serine and glutamic acid (Fig. 5). The sequence of this loop forming amino acids constitute an exception while mutation of the mdnA gene (Ziemert et al., 2008 and 2010).
To find optimized and novel microviridin variants, several libraries carrying modifications in the sequence of the MdnA core peptide were established. These libraries were generated by randomized oligonucleotide synthesis. Therefore a forward oligonucleotide displaying a part of conserved leader peptide sequence was built. Further a reverse oligonucleotide was created. The first part of this oligonucleotide is 20 bases, which overlap with the forward oligonucleotide and which is necessary for hybridization of both, forward and reverse, oligonucleotides. In the middle of the sequence modified bases were inserted. For cloning, the forward oligonucleotide starts with the blunt restricted site of SfoI. This results therein that the oligonucleotide can ligate in the SfoI restricted vector without previous digest of the oligonucleotide. The last part of the reverse oligonucleotides displays an AatII restriction site for cloning. Digestion with AatII results in sticky ends, thus wrong way round insertion of the oligonucleotide is impossible. Forward and reverse oligonucleotides were combined by performing a fill-in reaction. Because we designed several libraries with different diversity rates, the modified bases, which are inserted in the middle of the reverse primer, vary. All of our generated libraries contain mutations of the sequence of the MdnA core peptide at several sites, but not at the loop forming sites. Thus we are thinking that cyclization of microviridin happens accurately. A library with a diversity of 45,360 (focused library 1) and a second focused library showing a minor diversity of 810 (focused library 2) were designed. The included sequence modifications plus the diversity of our libraries are shown in figure 6. In our further studies we worked on focused library 2 primarily. In this library the nucleotide sequences encoding glycine residues were changed in GVK in all cases (Figure 6). GVK stands for guanine at the first position, adenine, cytosine or guanine at the second position and further thymine at the third position of the codon. These three possible codons encode for the amino acids alanine, aspartic acid and glycine. The nucleotide sequence N-terminal tyrosine residue was changed in THT describing thymine (first position), adenine, cytosine or thymine at the second position and thymine (third position). These codons encode isoleucine, leucine and phenylalanine. Additionally, the sequence of phenylalanine in the core peptide sequence was shift to NNK. NKK stands for one of the four nucleotides at the first position, and guanine or thymine at the second and third position. Consequently, this codon encodes the amino acids arginine, glycine, tryptophan, methionine, leucine, valine, serine, cysteine, isoleucine and phenylalanine.
To construct this library a fill-in reaction using the designed forward and reverse oligonucleotides was performed. Subsequently resulting randomized oligonucleotide was digested with the restriction enzyme AatII. In addition, the vector pUP089 was digested with the restriction enzymes AatII and SfoI. After ligation of fragments, randomized oligonucleotide and vector, the focused library 2 (ligation product) was transformed in chemocompetent E. coli XL1-Blue cells. This procedure was done several times and a library size of 1233 colonies was reached.
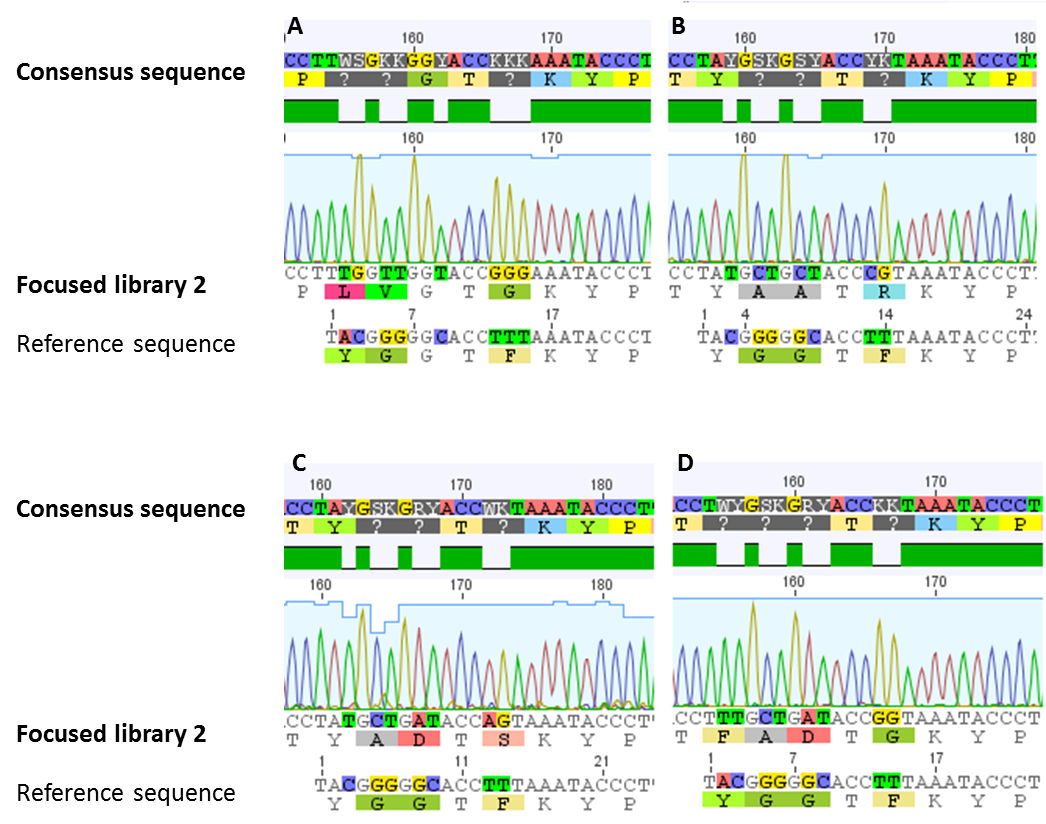
For confirmation of the focused library 2, sequencing analysis of a number of clones was performed. In all of our sequenced samples the requested modifications were established (figure 7). A sequence logo was generated for the 14 amino acid sized core peptide of MdnA. This sequence logo, which is illustrated in figure 8, indicates that the desired sites stay conserved. The others are modified.
The generated and modified focused library 2 should be used as basis for in vivo selection and phage display screening. Furthermore we purpose the objective to search for optimized and novel microviridins by using this library.
Expression Backbones
In a subtask of our work we wanted to construct auxiliary expression backbones with inducible promotors. In contrast to constitutive systems inducible systems only express the protein of interest after adding an inducer, which can be for example AHL or arabinose. We decided to construct IPTG- and arabinose-inducible systems. Therefore we amplified the promotor region and fused it to a reporter gene, which is constituted by YFP. Using this we have the option to verify the presence of the inducible promotor and also the function of the induction process by fluorescence. Our idea was to use the reporter gene as a placeholder. Via restriction enzyme digestion using the iGEM restriction enzyme sites you will be able to replace the reporter gene with your gene of interest. As vector backbone we made use of the pSB1A3, which has ampicillin resistance (Fig. 9). The constructs using pSB1K3 (kanamycin resistance) and pSB1C3 (chloramphenicol resistance) are still on the anvil.
We successfully tested the IPTG- and arabinose-inducible system (Fig 10). Using fluorescence microscopy we were able to detect the YFP-expression after an induction time of approximately 1.5 hrs. To prove the hypothesis we opposed an induced sample with a non-induced control for each promotor. By use of brightfield combined with differential interference contrast microscopy we traced the E. coli cells and switched then to the YFP detecting channel to investigate the fluorescence of these cells.
We also tested the inducible systems by using fluorescence spectroscopy. For this experiment we induced both systems at a time when the cultures were located in phase of exponential growth. After inducing, the cultures were analyzed by fluorescence spectroscopy. In this analyze the cells were excited by 500 nm. The resultant emission was measured in a spectrum between 510 and 580 nm. (Fig 11;12)
The lacking of the LacI gene on the vector is the reason for expression of YFP in both controls. For further research in this field it is necessary to clone the LacI gene inside the constructed vector.
References
Ziemert, N., Ishida, K., Liaimer, A., Hertweck, C. & Dittmann, E. (2008). Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angewandte Chemie (International ed. in English) 47, 7756-9
Ziemert, N., Ishida, K., Weiz, A., Hertweck, C. & Dittmann, E. (2010). Exploiting the natural diversity of microviridin gene clusters for discovery of novel tricyclic depsipeptides. Applied and environmental microbiology 76, 3568-74
 "
"