Team:Potsdam Bioware/Labjournal/September part 2
From 2011.igem.org
| (7 intermediate revisions not shown) | |||
| Line 629: | Line 629: | ||
<br> | <br> | ||
| - | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> Colony PCR of ''E. coli'' XL1 blue transformed with ligation products of mutated TEV protease, mutated | + | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;"> Colony PCR of ''E. coli'' XL1 blue transformed with ligation products of mutated TEV protease, mutated 14_3C protease, AraC form pBAD_iGEM_express, pJC354_ssTorA_XhoI_CS-14_3C_NheI_blaFL and pJC354_ssTorA_XhoI_CS-TEV_NheI_blaFL and analytical AG (1.5%)</h3> |
<b>Investigators:</b> Stefan, Sascha, Sebastian<br> | <b>Investigators:</b> Stefan, Sascha, Sebastian<br> | ||
| Line 677: | Line 677: | ||
* TEV1-TEV3 (clones with ligation product of TEV protease, AraC and pJC354_ssTorA_XhoI_CS-TEV_NheI_blaFL, approx. 1,6 kbp) | * TEV1-TEV3 (clones with ligation product of TEV protease, AraC and pJC354_ssTorA_XhoI_CS-TEV_NheI_blaFL, approx. 1,6 kbp) | ||
| - | * TEVbb1+TEVbb2 (clones with ligation product of | + | * TEVbb1+TEVbb2 (clones with ligation product of 14_3C protease, AraC and pJC354_ssTorA_XhoI_CS-TEV_NheI_blaFL, approx. 1,6 kbp) |
| - | * | + | * 14_3C1-14_3C5 (clones with ligation product of 14_3C protease, AraC and pJC354_ssTorA_XhoI_CS-14_3C_NheI_blaFL, approx. 1,5 kbp) |
| - | * | + | * 14_3Cbb1+14_3Cbb2 (clones with ligation product of TEV protease, AraC and pJC354_ssTorA_XhoI_CS-14_3C_NheI_blaFL, approx. 1,5 kbp) |
<b>Primer:</b><br> | <b>Primer:</b><br> | ||
| Line 691: | Line 691: | ||
*Primer 2 - r_TEV_ACCAGC | *Primer 2 - r_TEV_ACCAGC | ||
| - | Primer for all clones containing | + | Primer for all clones containing 14_3C protease: |
*Primer 1 - f_AraC_iGEM_HindIII | *Primer 1 - f_AraC_iGEM_HindIII | ||
| - | *Primer 2 - | + | *Primer 2 - r_14_3C_tm_XbaI280_A>T |
<br> | <br> | ||
| Line 703: | Line 703: | ||
<br> | <br> | ||
| - | Clones | + | Clones 14_3C5 (pUP_SG18_ssTorA_CS-14_3C_blaFL_AraC-14_3C5) and Tevbb1 (pUP_SG19_ssTorA_CS-TEV_blaFL_AraC-14_3C1) showed bands with the right size. All other clones showed no visible bands. Precultures are going to be cultivated over night at 37°C 750 rpm in thermo block. PCR will be repeated tomorrow.<br> |
100µl of the positive clones are diluted in 5 ml LB media (with chloramphenicol) and incubated over night at 37°C 250 rpm for mini prep of the plasmid DNA.<br> | 100µl of the positive clones are diluted in 5 ml LB media (with chloramphenicol) and incubated over night at 37°C 250 rpm for mini prep of the plasmid DNA.<br> | ||
| Line 2,851: | Line 2,851: | ||
<br> | <br> | ||
| - | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;">Miniprep of | + | <h3 style="background-color: rgb(100, 150, 100); font-weight: bold;">Miniprep of 14_3C2,3,4 and TEv 2,3,4</h3> |
<b>For better understanding of described experiment see also: [[http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx]]</b><br> | <b>For better understanding of described experiment see also: [[http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx]]</b><br> | ||
| Line 3,187: | Line 3,187: | ||
<br> | <br> | ||
<b>Results:</b><br> | <b>Results:</b><br> | ||
| - | [[file:UP_emission_Ara.png]] | + | [[file:UP_emission_Ara.png||400px]] |
| - | [[file:UP_emission_lac.png]] | + | [[file:UP_emission_lac.png||400px]] |
<br> | <br> | ||
<b>further tasks:</b><br> | <b>further tasks:</b><br> | ||
*analyze by fluorescence microscopy | *analyze by fluorescence microscopy | ||
| + | |||
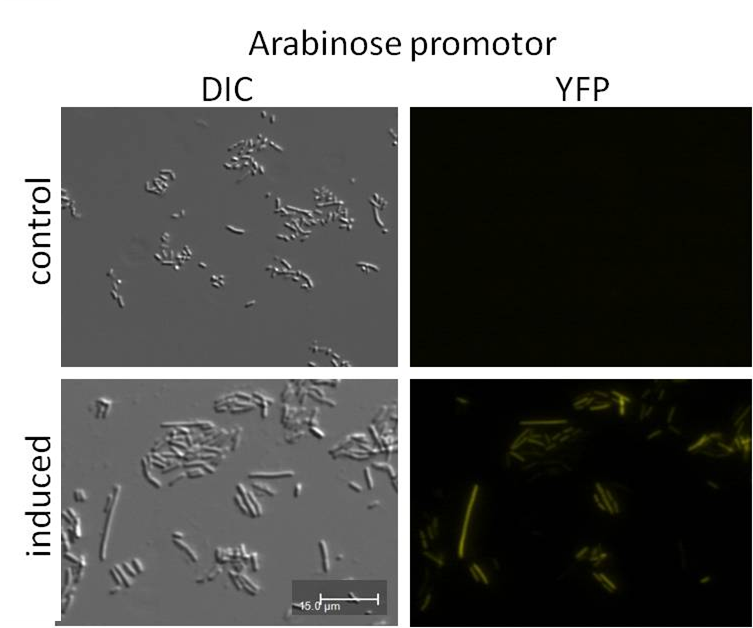
| + | <h3 style="background-color: rgb(254, 122, 122); font-weight: bold;">Analysis of psB1A3_Ava_YFP and psB1A3_IPTG_YFP by fluorescence microscopy. | ||
| + | </h3> | ||
| + | |||
| + | <b>Investigators:</b> Katharina<br> | ||
| + | |||
| + | <br> | ||
| + | <b>Materials:</b><br> | ||
| + | *psB1A3_Ava_YFP over night culture | ||
| + | *psB1A3_IPTG_YFP over night culture | ||
| + | |||
| + | <br> | ||
| + | <b>Methods:</b><br> | ||
| + | *1 ml culture were transfered in new media | ||
| + | *observed growing by measuring OD(600nm) | ||
| + | *inducing of promotor by 5 mM Ara or 1 mM IPTG at a OD(600nm) by 0.4-0.5) | ||
| + | *measuring of samples by fluorescence microscopy (1h after inducing) | ||
| + | |||
| + | <br> | ||
| + | <b>Results:</b><br> | ||
| + | |||
| + | |||
| + | [[file:UP Expression Backbones Ara.png]] | ||
| + | |||
| + | |||
| + | <h3 style="background-color: rgb(254, 122, 122); font-weight: bold;">Sending of BioBricks to MIT | ||
| + | </h3> | ||
| + | |||
| + | <b>Investigators:</b> Nicole, Nadine, Sebastian<br> | ||
| + | |||
| + | '''BioBricks''' | ||
| + | {| class="wikitable" border="1" | ||
| + | |||
| + | |- | ||
| + | |||
| + | ! Label number !! BioBrick Nickname !! Tube label !! c [ng/µl] | ||
| + | |||
| + | |- | ||
| + | |||
| + | | BBa_K627000 || mdnED || 1 || 25 | ||
| + | |||
| + | |- | ||
| + | |||
| + | | BBa_K627001 || mdnA || 2 || 25 | ||
| + | |||
| + | |- | ||
| + | |||
| + | | BBa_K627002 || mdnB || 3 || 25 | ||
| + | |||
| + | |- | ||
| + | |||
| + | | BBa_K627003 || mdnC || 4 || 25 | ||
| + | |||
| + | |- | ||
| + | |||
| + | | BBa_K627004 || mdnD || 5 || 25 | ||
| + | |||
| + | |- | ||
| + | |||
| + | | BBa_K627005 || mdnE || 6 || 25 | ||
| + | |||
| + | |- | ||
| + | |||
| + | | BBa_K627006 || mdnA c-myc gene III || 7 || 25 | ||
| + | |||
| + | |- | ||
| + | |||
| + | | BBa_K627007 || c-myc gene III || 8 || 25 | ||
| + | |||
| + | |- | ||
| + | |||
| + | | BBa_K627008 || AraC TEV protease 1 || 9 || 25 | ||
| + | |||
| + | |- | ||
| + | |||
| + | | BBa_K627009 || AraC TEV protease 2 || 10 || 25 | ||
| + | |||
| + | |- | ||
| + | |||
| + | | BBa_K627010 || AraC TEV protease 3 || 11 || 25 | ||
| + | |||
| + | |- | ||
| + | |||
| + | | BBa_K627011 || AraC 14_3C protease || 12 || 25 | ||
| + | |||
| + | |- | ||
| + | |||
| + | | BBa_K627012 || ssTorA CS-TEV BlaFL || 13 || 25 | ||
| + | |||
| + | |- | ||
| + | |||
| + | | BBa_K627013 || ssTorA CS-14_3C BlaFL || 14 || 25 | ||
| + | |||
| + | |- | ||
| + | |||
| + | | BBa_K627014 || A3_Ara_YFP clone 1 || 15 || 25 | ||
| + | |||
| + | |- | ||
| + | |||
| + | | BBa_K627015 || A3_lac_YFP clone 2 || 16 || 25 | ||
| + | |} | ||
| + | <br> | ||
Latest revision as of 00:26, 22 September 2011
92th Labday 2011-09-11
miniprep of pSB1C3+mdnABCDE with and without T7 promotor
Time: 2011-09-11
Investigators: Niels, Jessica, Katharina
Materials:
- 6 overnight cultures
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- Protocol for high-copy plasmids
- elution with 50 µl H2O
- measuring concentration with NanoDrop:
| Sample | concentration in ng/µl |
|---|---|
| mdnABCDE with T7-promotor clone 1 | 50.6 |
| mdnABCDE with T7-promotor clone 2 | 10.6 |
| mdnABCDE without T7-promotor clone 1 | 68.4 |
| mdnABCDE without T7-promotor clone 2 | 110.2 |
| mdnABCDE without T7-promotor clone 3 | 108.0 |
| mdnABCDE without T7-promotor clone 4 | 22.3 |
| mdnABCDE without T7-promotor clone 5(III) | 504.0 |
| mdnABCDE without T7-promotor clone 6 (IV) | 212.6 |
miniprep of mdnA foc2 part 2
Time: 2011-09-11, 9:00
Investigators: Jessica, Niels, Katharina
Materials:
- 1 overnight culture
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- Protocol for high-copy plasmids
- elution with 50 µl H2O
- measuring concentration with NanoDrop:
| Sample | concentration in ng/µl |
|---|---|
| mdnA foc2 | 155.7 |
Further Tasks:
- give it to the screening group
PCR mdnABCDE with Long PCR Enzyme mix from Fermentas
Time: 2011-09-11
Investigators: Jessica, Katharina
Aim: generate BioBrick of mdnABCDE, amplify mdnABCDE for ligation in pSB1C3
Materials
- vector: pARW089 (8,6 ng/µl), pARW071 (6,3 ng/µl)
- DNA polymerases: Long PCR Enzyme Mix (Fermentas)
- Template DNA: pARW071 (vector)
- dNTPs
- water
- Primer:
| # | Primer |
|---|---|
| 84 | pf_mdnABCDE89_EcoRI_NotI_XbaI (froward for pARW071 and pARW089) |
| 82 | r_mdnABCDE_iGEM (reverse for all) |
| 83 | pf_mdnABCDE89+T7_EcoRI_NotI_XbaI (froward for pARW089, generating mdnABCDE+T7) |
Protocol:
- mdnABCDE
- 2 µl vector
- 1 µl dNTPs (10 mM)
- 3 µl forward Primer
- 3 µl reverse Primer
- 0.3 µl Polymerase
- 5 µl Buffer Fermentas Long Enzyme Mix (+MgCl2)
- 35.3 µl water
- total volume: 50 µl
2. PCR program
- IGLONG3
- first steps: 10x
- second steps: 25x
| Step | Temperature | Time |
|---|---|---|
| Hot Start | 94°C | Hold |
| Initial denaturation | 94°C | 180 sec |
| Denaturation | 94°C | 15 s |
| Annealing | 59 | 30 s |
| Elongation | 68°C | 324 s |
| DenaturationII | 94°C | 15 s |
| AnnealingII | 68°C | 30 s |
| ElongationII | 68°C | 324 s + 2 s per each cycle s |
| Final Elongation | 68°C | 600 s |
Results:
Further tasks:
- PCR purification
- digest
- ligation
PCR of mdnA with C-terminal myc tag (mycC)
Investigators: Katharina, Jessica
Time: 2011-09-11, 14:00
Material:
- pARW089 (~8 ng/µl)
- dNTPs
- primer 23, 24
- HF Phusion Buffer 5x
- Phusion Polymerase
Method:
- 1 µl pARW089
- 1 µl dNTPs (10 mM each)
- 2.5 µl forward primer (10µM)
- 2.5 µl reverse primer (10µM)
- 10 µl HF Phusion Buffer 5x
- 0.5 µl Phusion Polymerase
- 32.5 µl water
- total 50 µl
- programm IGBIO2
| Step | Temperature | Time | |
|---|---|---|---|
| Hot Start | 98°C | Hold | |
| Initial denaturation | 98°C | 30 sec | |
| Denaturation | 98°C | 10 s | 10x |
| Annealing | 51°C | 20 s | |
| Extension | 72°C | 20 s | |
| Denaturation | 98°C | 10 s | 20x |
| Annealing | 72°C | 20 s | |
| Extension | 72°C | 20 s | |
| Final extension | 72°C | 10 min | |
| 4°C | Hold |
Result:
- PCR product mycN (stored in ?)
Further Tasks:
- restriction enzyme digestion
- ligation
- transformation
Restriction enzyme digestion of mycN and pSB1C3
Investigators: Jessica, Katharina
Time: 2011-09-11
Material:
- purified PCR products
- mycN (40.0 ng/µl)
- NEB Buffer 4
- XbaI
- PstI
- BSA
- digestion of mycN
- 20 µl DNA
- 3 µl Buffer 4
- 1.5 µl XbaI
- 2.0 µl PstI
- 0.3 µl BSA
- 3.2 µl water
- digestion of pSB1C3
- 6 µl DNA (235.0 ng/µl (2011-06-22))
- 3 µl Buffer 4
- 1.5 µl XbaI
- 2.0 µl PstI
- 0.3 µl BSA
- 11.5 µl water
- concentration after digestion
- mycN: 12.2
- pSB1C3: 12.1
PCR Purification of mdnA+mycC, Lib 2 for Phage Display and digested mdnA+mycN
Time: 2011-09-11
Investigators: Niels, Jessica, Katharina
Materials
- Macherey-Nagel Nucleo Spin Extract II
- elution in 50 µl
- concentration
- mdnA+mycC: 42.6 ng/µl
- Lib 2 for Phage Display: 27.9 ng/µl
- digested mdnA+mycN: 12.2 ng/µl
Gel electrophoresis of pARWIII and pSB1C3
Investigators: Niels, Jessica, Katharina
Time: 2011-09-11
Material:
- 1% agarose gel
- 2 µl Gel Red
- Fermentas DNA Ladder Mix
| lane | Sample | Volume in µl |
| M | Gene Ruler DNA Ladder Mix (1:10) | 12 |
| 1 | pARWIII | 30 + 6 loading dye |
| 2 | pSB1C3 (1) | 30 + 6 loading dye |
| 3 | pSB1C3 (2) | 30 + 6 loading dye |
- fragments were excised and purified using the Macherey-Nagel Nucleo Spin extract II Kit
- concentration after purification
- pARWII: 3.9 ng/µl
- pSB1C3 (1): 8.4 ng/µl
- pSB1C3 (2): 12.1 ng/µl
Restriction enzyme digestion of pSB1C3 for control
Investigators: Katharina, Jessica, Niels, Steffi
Time: 2011-09-11
Material:
pSB1C3(2011-06-22) :
- concentration: 235,0 ng/µl
digestion of pSB1C3 with Pvu II:
- 2 µl DNA (235.0 ng/µl (2011-06-22))
- 1 µl 10xBuffer
- 1 µl PvuII
- 6 µl water
- total volume 10 µl
- 37°C over night
further tast
electrophoretic separation
Ligation of pARWIII + Lib-2 for Phage Display and pSB1C3 + mdnAmycN
Time: 2011-09-11
Investigators: Steffi, Niels, Katharina, Jessica
Material
- purified pARWIII (vector) (2011-09-11)
- purified Lib-2 for Phage Display (insert) (2011-09-11)
- purified pSB1C3 (2011-09-11)
- purified mdnA + mycN
Method
- ligation of pARWIII + Lib2 for Phage Display
- 1 µl 10x Buffer
- 1 µl T4 Ligase
- 7 µl vector
- 1 µl insert
- 1h @ RT
- ligation of pSB1C3 + mdnAmycC
- 1 µl 10x Buffer
- 1 µl T4 Ligase
- 7 µl vector
- 1 µl insert
- 1h @ RT
- control for each ligation: taking water instead of insert
Further tasks:
- transformation
Testing of expression backbones
Time: 2011-09-11
Investigators: Niels, Katharina, Jessica
Material
- overnight cultures of pSB1K3_Ara_CFP and pSB1K3_Lac_CFP
Results:
- cells didn't grow properly
Further tasks:
- repeat with new overnight cultures
Preparing new overnight cultures of expression backbones
Time: 2011-09-11
Investigators: Niels, Katharina, Jessica
Material
- glycerol stocks of pSB1K3_Ara_CFP and pSB1K3_Lac_CFP
- stocks weren't prepared correctly, so
- plates of pSB1A3_YFP_Ara, pSB1A3_YFP_Lac, pSB1K3_CFP_Ara, pSB1K3_CFP_Lac, pSB1K3_YFP_Lac
Further tasks:
- expression test with pSB1K3_Ara_CFP and pSB1K3_Lac_CFP
- glycerol stocks of all 5 cultures
Transformation of pARWIII + Lib-2 for Phage Display
Time: 2011-09-11
Investigators:Niels, Jessica, Katharina, Steffi
Aim: Transformation of E. coli cells with ligation of pARWIII + Lib-2 for Phage Display
Materials:
- competent E. coli cells (XL1-Blue, 2011-08-29)
- ligation products: pARWIII + Lib-2 for Phage Display
Method:
- addition of 2 µl ligation reaction to cells (XL1-blue) in 1.5 ml Eppi,
- incubation 20 min on ice,
- heat shock 90 sec at 42°C,
- incubation 10 min on ice,
- addition of 750 µl LB medium,
- incubation at 37 °C shaking(750 rpm) for 60 min,
- centrifuge 30 min at 2000 x g
- discard the supernatant (till ~100µl media)
- plating on LB plate with appropriate antibiotic (Amp)
- storage over night at 37°C
Further tasks:
- counting colonies
- wash colonies from plates
- o.n. cultures and miniprep
Colony PCR of E. coli XL1 blue transformed with ligation products of mutated TEV protease, mutated 14_3C protease, AraC form pBAD_iGEM_express, pJC354_ssTorA_XhoI_CS-14_3C_NheI_blaFL and pJC354_ssTorA_XhoI_CS-TEV_NheI_blaFL and analytical AG (1.5%)
Investigators: Stefan, Sascha, Sebastian
Aim: checking for positive clones for further tasks
Methode:
- picked clones where incubated over night at 4°C in 1 ml LB media
- 2 µl were used as template for PCR
- 2 µl were used as template for PCR
- 5 µl Buffer S with 25 mM MgCl2 (purchased by Genaxxon)
- 2,5 µl each Primer (10 mM, see list below)
- 1 µl dNTP (each 10 mM)
- 2 µl Templates
- 1 µl Taq Polymerase (purchased by Genaxxon)
- 2 µl 25 mM MgCl2 (purchased by Genaxxon)
- 34 µl H2O
- Total volume: 50 µl
PCR Program:
Initial denat = 5min 98°C
25x
denat: 1min 98°C
anneal: 1min 45sec 55°C
extend: 1min 45sec 72°C
final extend: 10min 72°C
Templates:
- TEV1-TEV3 (clones with ligation product of TEV protease, AraC and pJC354_ssTorA_XhoI_CS-TEV_NheI_blaFL, approx. 1,6 kbp)
- TEVbb1+TEVbb2 (clones with ligation product of 14_3C protease, AraC and pJC354_ssTorA_XhoI_CS-TEV_NheI_blaFL, approx. 1,6 kbp)
- 14_3C1-14_3C5 (clones with ligation product of 14_3C protease, AraC and pJC354_ssTorA_XhoI_CS-14_3C_NheI_blaFL, approx. 1,5 kbp)
- 14_3Cbb1+14_3Cbb2 (clones with ligation product of TEV protease, AraC and pJC354_ssTorA_XhoI_CS-14_3C_NheI_blaFL, approx. 1,5 kbp)
Primer:
Primer for all clones containing TEV protease:
- Primer 1 - f_AraC_iGEM_HindIII
- Primer 2 - r_TEV_ACCAGC
Primer for all clones containing 14_3C protease:
- Primer 1 - f_AraC_iGEM_HindIII
- Primer 2 - r_14_3C_tm_XbaI280_A>T
Results:
Clones 14_3C5 (pUP_SG18_ssTorA_CS-14_3C_blaFL_AraC-14_3C5) and Tevbb1 (pUP_SG19_ssTorA_CS-TEV_blaFL_AraC-14_3C1) showed bands with the right size. All other clones showed no visible bands. Precultures are going to be cultivated over night at 37°C 750 rpm in thermo block. PCR will be repeated tomorrow.
100µl of the positive clones are diluted in 5 ml LB media (with chloramphenicol) and incubated over night at 37°C 250 rpm for mini prep of the plasmid DNA.
Picture of GelDoc will follow as soon as possible
Further tasks:
- overnight culture for plasmid preparation of pUP_SG18... and pUP_SG19...(sequencing and preparing biobricks)
- creating glycerol stock cultures form all positive clones (XL1 blue transformed with pUP_SG18... and XL1 blue transformed with pUP_SG19)
- if pUP_SG19... doesnt show any unwanted mutations, this clones needs to be digested with XbaI and PstI and ligated with digested pUP_SG13... (also digested with XbaI and PstI)
- survival screening with pUP_SG18... and pUP_SG19... after sequencing without any unwanted mutations
93th Labday 2011-09-12
Preparation of samples for HPLC analysis part II
Time: 2011-09-12
Investigators: Jessica, Katharina
Material:
- methanol
- PALL Syringe Filter
- HPLC vial
Method:
- the pellet obtained after speedvac was resuspended in 200µl of 80% methanol
- centrifugation for 3 min @ 13000 rpm
- samples were filtered using a small PALL Syringe Filter
- the filtered sampled was put into a HPLC vial
- performance of HPLC
competent cells - E. coli XL1 blue
Investigator: Katharina, Niels, Steffi
Aim: produce competent cells
Materials/Methods:
| TFB I | 1000ml | 200ml |
| 100mM Rubidium Chloride | 12.1 | 2.42g |
| 30mM Potassium Acetate | 2.944 | 0.59g |
| 10mM Calcium Chloride | 1.47 | 0.29g |
| 15% w/v Glycerol (87%) | 150 | 34.5g |
Adjust pH to 5.8 with acetic acid
Filter sterilize the solution
| TFB II | 500ml | 100ml |
| 50mM Rubidium Chloride | 0.6 | 0.121g |
| 10mM MOPS | 1.05 | 0.210g |
| 75mM Calcium Chloride | 5.51 | 1.100g |
| 15% w/v Glycerol (87%) | 75 | 17.24g |
Adjust pH to 7.0 with KOH
Filter sterilize the solution
Work always sterile and cold and speedy!
- All volumes deal with the common cellline!
- Prepare 80 Eppis (1,5?l)
- get liquid nitrogen
- prepare 5 ml LB-Medium with the specific antibiotic (for XL1-blue: Tet), inoculate and incubate over night
- prepare 200 ml LB-Medium with the specific antibiotic, inoculate with 2 ml of the over-night-culture
- grow while shaking at 37°C, 190 rpm to an OD600 at 0,4-0,6
- keep cell suspension in sterile falcons (50 ml) 10 min on ice, then centrifuge for 5 min, 4°C, 4000 rpm
- discard supernatant, carefully resuspend on ice with 40 ml icecold TFB I and keep 10 min on ice
- centrifuge for 5 min, 4°C, 4000 rpm
- discard supernatant, carefully resuspend pellet in 8 ml TFB II
- aliquot in Eppis: 50?l per tube and store immediately at liquid nitrogen and afterwards at -80 °C
Results:
- 80 tubes XL1 blue for transformation (50 µl competent cells at -80°C)
Further tasks:
check by transformation and check the resistance on agar plates with different antibiotics
check resistance of competent cells - E. coli XL1 blue
Investigator: Katharina, Niels, Steffi
Aim: check resistance of competent XL1 blue-cells on agar plates with different antibiotics
Materials/Methods:
- competent XL1 blue-cells from 2011-09-12 (Katharina, Niels, Steffi)
- LB-plates with Tetracycline, Chloramphenicol, Kanamycine, Ampicillin
- plating 50 µl on agar plates
- incubate at 37°C over night
Further tasks:
control agar plates
Digest of PCR products mycC, mdnABCDE 71, 89, T7 and pSB1C3
Time: 2011-09-12
Investigators: Jessica, Nicole, Nadja
Materials
- PCR products:
- mycC
- mdnABCDE 71
- mdnABCDE 89
- mdnABCDE T7
- pSB1C3
- EcoRI, SpeI, BSA, NEB Buffer 4
Reaction mix
- for PCR products:
- 10 µl PCR product
- 1.5 µl EcoRI
- 1.5 µl SpeI
- 2 µl Buffer 4
- 0.2 µl BSA
- 4.8 µl water
- for pSB1C3:
- 14.8 µl pSB1C3
- 1.5 µl EcoRI
- 1.5 µl SpeI
- 2 µl Buffer 4
- 0.2 µl BSA
Output
- digests:
- mycC
- mdnABCDE 71 a
- mdnABCDE 71 b
- mdnABCDE 89 a
- mdnABCDE 89 b
- mdnABCDE T7 a
- mdnABCDE T7 b
- pSB1C3
- pSB1C3
Further tasks
- gel electrophoresis
Purification of mycC, digested mdnABCDE 71-a/b, 89-a/b, T7-a/b and digested pSB1C3 from today
Time: 2011-09-12
Investigators: Jessica, Nicole, Nadja
Materials
- Macherey-Nagel Nucleo Spin Extract II
- elution in 35 µl H2O (of mycC and mdnABCDE 71-a/b, 89-a/b, T7-a/b)
- elution in 50 µl NE eluation buffer ( of pSB1C3)
- concentration
- mycC: 23,9 ng/µl
- mdnABCDE 71 a: 135,2 ng/µl
- mdnABCDE 71 b: 48,7 ng/µl
- mdnABCDE 89 a: 26,8 ng/µl
- mdnABCDE 89 b: 9,9 ng/µl
- mdnABCDE T7 a: 89,0 ng/µl
- mdnABCDE T7 b: 120,2 ng/µl
- pSB1C3: 32,5 ng/µl
- pSB1C3: 32,5 ng/µl
Further tasks
- ligation
Ligation of mycC and digested mdnABCDE 71-a/b, 89-a/b, T7-a/b with digested pSB1C3 from today
Time: 2011-09-12
Investigators: Jessica, Nicole, Nadja
Materials
- pSB1C3
- mycC and digested mdnABCDE 71-a/b, 89-a/b, T7-a/b
Protocol
Components
| Component | Molar Ratio | Concentration in ng/ µl | Length in bp | volume |
| Backbone pSB1C3 | 1 | 32,5 | 2390 | fill up to 8µl |
| Insert: mdnABCDE 71-a | 3 | 135,2 | 6500 | 5 |
| Insert: mdnABCDE 71-b | 3 | 48,7 | 6500 | 7 |
| Insert: mdnABCDE 89-a | 3 | 28,8 | 6500 | 7 |
| Insert: mdnABCDE 89-b | 3 | 9,9 | 6500 | 7 |
| Insert: mdnABCDE T7-a | 3 | 89,0 | 6500 | 6 |
| Insert: mdnABCDE T7-b | 3 | 120,2 | 6500 | 6 |
| Insert: mycC | 3 | 23,9 | 220 | 1 |
Ligation mix
- * * for a:
- 1µl T4 ligase
- 1µl T4 buffer
- fill up with water
- total volume 10 µl
- at 14 C over night
- * * for b:
- 1µl quck ligase
- 10µl quick ligase buffer
- fill up with water
- total volume 20 µl
- at 14 C over night
Further task:
- Transformation
ELISA with purified phages
Investigators: Sandrina
Aim: control if mdnA-myc-geneIII will be expressed on the phage
Method/Materials:
- ELISA plate coated with 5 µg/ml anti-c-myc antibody in phosohate buffer (pH 7,4), volume 100 µl/ well
- incubate for 4 h at room temperature
- blocking in 2 % milk powder in TBS (300 µl/ well)
- incubate for 2 h at room temperature
- add 100 µl phages, produced in XL1-blu cells and in ER2738 cells diluted 1:5 in TBS-T (0,005%)
- incubate shaking for 60 min at room temperature
- wash 5 x with TBS-T (0,05%)
- add anti-gene-8-antibody (HRP coupled)
- incubate shaking for 60 min
- wash 5 x with TBS-T
- put substrate on the samples
- measure in plate photometer
- reference: helper phages
Resluts:
- x-axis: 1) absorption negative control (helper phages) 2) absorption of phages produced in XL1-blue cells
- conclusion: geneIII-myc-mdnA-fusion gene is expressed on the surface of phages produced in XL1-blue cells
- at wells incubated with phages produced in ER2738 cells no significant signals were observed
Further tasks:
Phage Display
Send clones 13, 14 (mdnA in pSB) and 7,8 (geneIII in pSB) for sequencing
Investigators: Sandrina
Aim:control ligation of mdnA and geneIII into pSB1C3 for generating biobricks
Method/Materials:
- Primer: VR
- 20 µl with 70 ng/µl DNA
Further tasks:
check alignment
94th Labday 2011-09-13
Preparation of samples for HPLC analysis
Time: 2011-09-13, 8:00
Investigators: Nicole, Jessica, Nadine, Katharina
Aim: Isolation of Microviridin from E. coli
Material:
- ON cultures (XL1-blue):
- pSB1C3+mdnABCDE klon 5 (cm resistance)
- pARW089 (kan resistance)
- pARW071 (kan resistance)
- LB medium
- kanamycin
- chloramphenicol
- induction solution (content???)
- 100% methanol
- 5& methanol
- sterile water
- Sep-Pak Cartridges
- speed vac
Method:
- prepare main cultures:
- 150 ml LB + 150 µl appropriate antibiotics
- add 1.5 ml ON culture
- add 150 µl induction solution (time: 8:30)
- incubation at 30°C and 200 rpm to an OD of ???
- incubation at 30°C and 200 rpm to an OD of ???
- harvesting and lysis
- pellet was resuspended in 5 ml of sterile water
- the resuspended sample was sonicated for 5 min (3 sec on, 3 sec off)
- centrifugation for 10 min @ 11000 rpm
- microviridin isolation
- supernatant was transfered to Sep-Pak Plus C18 Cartridges, that were equilibrated with 2 ml of 100% methanol
- cartridges were washed with 2 ml water
- samples were load
- cartridges were washed with 2 ml of 5% methanol
- samples were eluted with 2 ml of 100% methanol
- samples were put in a speedvac so that the methanol can evaporate
Note:
- procedure was stopped because we had positive results from first HPLC
Conclusions:
- system has not to be induced (only fosmids have to be induced)
- HPLC will be repeated w/ remaining sample (2011-9-12), fractions will be collected and analyzed by Mass Spectrometry
HPLC w/ samples from 2011-9-12 (repetition)
Time: 2011-09-13, 8:00
Investigators: Nicole, Nadine, Katharina, Vanessa, Nadja
Aim: this time: collect fraction during the run for MS
Material:
- microviridin samples (from pARW089 and pARW071) from 2011-9-12
- HPLC (Shimadzu)
HPLC program:
Results:
Conclusions:
Test Expression Backbones: pSB1K3_Ara/Lac_CFP
Time: 2011-09-13
Investigators:
Niels
Aim:
prove of promotors (Lac/Ara)
Materials
- pSB1K3_Lac_CFP culture
- psB1K3_Ara_CFP culture
Methods
- cells from overnight cultre (2011-09-12)were transfer in new media (50ml LB + 50µl Kan (50 mg/ml))
- start cell grow in fresh media
- inducing of Ara at 0,6 (OD600)
- after inducing by IPTG/ARA - samples where measured for 2h every 15 min
- results negativ
further task
- prove of fluorescence by microscope
Expression Backbones: prepare samples for fluorescence microscope
Time: 2011-09-13
Investigators: Nicole,Vanessa, Jessica, Nadine, Nadja
Aim:
- plate clones on induced and non-induced plates
- also: liquid culture
Materials
- pSB1K3_Lac_CFP culture
- psB1K3_Ara_CFP culture
- pSB1A3_Lac_YFP culture
- psB1A3_Ara_YFP culture
- LB-Agar
- kanamycin
- ampicilin
- arabinose
- IPTG
Digest of pSB1C3 for mdnABCDE
Time: 2011-9-7, 11:00
Investigators: Nadine, Nicole
Aim: generate a BioBrick of mdnABCDE, digest for ligation in pSB1C3
Materials:
- BBa_K404304_pSB1C3 (#3), 284.6 ng/µl
- EcoRI, SpeI, PstI
- Buffer 4
- BSA
- H2O
Protocol:
- vector (2 batches: batch 1 w/ SpeI and batch 2 w/pstI):
- 5 µl vector
- 1 µl EcoRI
- 1 µl SpeI/PstI
- 2 µl Buffer 4
- 0.2 µl BSA
- 10.8 µl H2O
- total volume: 20µl
- incubation 3 hrs at 37°C (start: 9:15)
Output:
Further tasks:
- agarose gel
- purification
- ligation
Transformation of XL1 with mdnABC and a control with H2O
Time: 2011-09-14; 23:00
Investigators: Nadine, Nicole, Jessica, Nadja
Materials:
- XL1-Blue
- ligation product: pSB1C3+mdnABC, Jessica, Nadja, 28.08.2011 resistance: cm
- LB-Agar plates
Method:
- ligation products:
- addition of 2 µl ligation reaction to cells (XL1-blue) in 1.5 ml Eppi,
- incubation 30 min on ice,
- heat shock 60 sec at 42°C,
- incubation 5 min on ice,
- addition of 750 µl LB medium,
- incubation at 37 °C shaking for 60 min,
- plating on LB medium with appropriate antibiotic (20 ml Agar and 20 µl antibiotic (see above) per plate)
- storage over night at 37°C (start: 15:00)
Further tasks:
- Picking clones for overnight culture
- Producing glycerol stocks
Overnight cultures of pARW071_mdnB and pARW089_mdnB
Time: 2011-09-07, 00:00
Investigators: Nadine ,Nicole, Jessica; Nadja
Aim: For characterization via Western blot
Materials
- LB medium with Amp
- glycerol stocks of of pARW071_mdnB and pARW089_mdnB
Method:
- inoculation of 10 ml LB medium with antibiotic
- overnight at 37°C
Further tasks:
- testing expression
Digestion set
Time: 2011-09-15, 00:00
Investigators: Nadine ,Nicole, Jessica; Nadja
Aim: To build the Biobrick pSB1C3_mdnABCDE
Method:
Digest compositions
| sample | mdnE | mdnD | mdnDE | mdnABC |
| ingrediants | Volume in µl | Volume in µl | Volumein µl | Volume in µl |
| DNA | 5,3 | 5,0 | 2,0 | 1,5 |
| buffer 4 | 3 | 3 | 3 | 3 |
| BSA | 0,3 | 0,3 | 0,3 | 0,3 |
| H2O | 18,7 | 18,7 | 21,7 | 22,2 |
| PstI | 1,5 | 1,5 | 1,5 | 1,5 |
| XbaI | 1,5 | - | 1,5 | - |
| SpeI | - | 1,5 | - | 1,5 |
- 1,5 h at 37°C
Further tasks:
- Agarose gel
- gel extraction
- ligation
Digestion set verification on Agarose Gel, gel extraction and ligation
Time: 2011-09-15, 01:15-05:30
Investigators:Nadine, Nicole, Jessica, Nadja
Materials
- digested mdn_ACD
- digested mdn_E
- digested mdn_D
- digested mdn_DE
Production of one 0,8 % gel for mdnABC and mdnDE and one 1% for mdnE and mdnD
- 0,8 % gel: 0.4 g agarose in 50 ml 1x TAE buffer
- 1 % gel: 0.5 g agarose in 50 ml 1x TAE buffer
- Adding 2 µl gel red to the gel
Loading gels and running
1. 0,8 % gel
| lane | Sample | Volume in µl | Expected size in bp |
| M | Gene Ruler DNA Ladder Mix | 15µl (1:100) | |
| 1 | mdn_ABC | 20µl + 4ml 6x loading dye | 4819 |
| 2 | mdn_DE | 20µl + 4ml 6x loading dye | 2759 (including mdnDE), 2377 |
2. 1% gel
| lane | Sample | Volume in µl | Expected size in bp |
| M | Gene Ruler DNA Ladder Mix | 15µl (1:100) | |
| 1 | mdn_D | 20µl + 4ml 6x loading dye | 2934 |
| 2 | mdn_E | 20µl + 4ml 6x loading dye | 2394, 2070 (including mdnE) |
- 100V
- Ca. 1h
Gel extraction:
- using Nucleospin Extract II
- NOTE: accidently elution in collection tube
- second elution with 35 µl NE Buffer
Ligation:
| Backbone | Concentration in ng/ µl | Length in bp | volume | Insert | Concentration in ng/ µl | Length in bp | volume |
| pSB1C3_ABC | 4.4 | 4819 | 7.3 | ED | 5.7 | 2759 | 9.7 |
| pSB1C3_D | 8.2 | 2934 | 4.7 | E | 6.6 | 2070 | 12.3 |
Ligation mix
- 1 µl T4 ligase
- 2 µl T4 buffer
- insert and vector according to table
- total volume 20 µl
- at RT for 45 min
Further task:
- transformation
Repeated: Digestion set
Time: 2011-09-15, 4:30
Investigators: Nadine ,Nicole, Jessica; Nadja
Aim: To build the Biobrick pSB1C3_mdnABCDE
Method:
Digest compositions
| sample | mdnE | mdnD | mdnDE | mdnABC |
| ingrediants | Volume in µl | Volume in µl | Volumein µl | Volume in µl |
| DNA | 5,3 | 5,0 | 2,0 | 1,5 |
| buffer 4 | 3 | 3 | 3 | 3 |
| BSA | 0,3 | 0,3 | 0,3 | 0,3 |
| H2O | 18,7 | 18,7 | 21,7 | 22,2 |
| PstI | 1,5 | 1,5 | 1,5 | 1,5 |
| XbaI | 1,5 | - | 1,5 | - |
| SpeI | - | 1,5 | - | 1,5 |
- ? h at 37°C
Further tasks:
- Agarose gel
- gel extraction
- ligation
Check sequenced vector pSB1C3 conataining mdnA-myc-geneIII
Investigators: Sabine, Sandrina
Aim:control if biobrick is generated correctly
Method/Materials:
- genious
- make alignment
Results:
- three positive clones (pSB1C3 containing mdnA-myc-geneIII) fusion gene
95th Labday 2011-09-14
Repeated PCR of mdnA for cloning it into pSB1C3
Investigator: Sabine, Sandrina
Time: 2011-09-14,10:30-12:30
Aim:
- amplification of mdnA with iGEM-restriction sites
Primer:
- primer: pf_mdnA_iGEM_EheI and pr_mdnA_iGEM_AatII
Reaction Components:
- 2 µl Vector pARW089 (16 ng)
- 0,25 µl Taq Polymerase S (BioScience)
- 1 µl dNTPs
- 1 µl per primer
- 5 µl 10x PCR Buffer S
- 39,75 µl water
Further tasks:
- purification
- digestion
Digestion of PCR product mdnA for cloning into pSB1C3
Investigator: Sandrina, Sabine
Aim: cloning of mdnA into pSB1C3
Time: 12:30-14:00
Material/Method:
- 30 µl PCR product mdnA
- 1 µl restriction enzyme XbaI
- 1 µl restriction enzyme PstI
- 4 µl NEB 10x buffer 2
- 0,4 µl BSA
- 3,6 µl water
- 1,5 h, 37°C
Further Tasks:
- gel electrophoresis for purification
- ligation of mdnA with pSB1C3
Ligation of mdnA with pSB1C3 to generate mdnA as a biobrick
Investigator: Sandrina, Sabine
Time: 2011-09-14, 14:00-15:00
Material/Method:
- 8,9 µl mdnA (4 ng/µl)
- 8,1 µl pSB1C3 (17 ng/µl)
- 2 µl 10x T4 ligase buffer 3
- 1 µl T4 ligase
- 1,4 µl water
- 1 h, room temperature
Further Tasks:
- Transformation in XL1-blue cells
Transformation of pSB1C3 containing mdnA in E. coli
Investigator: Sandrina, Sabine
Time: 2011-09-14, 15:00-16:30
Aim:amplification of vectors
Method:
- addition of 4 µl ligation reaction to XL1-blue cells
- incubation 20 min on ice,
- heat shock 45 sec at 42°C,
- incubation 2 min on ice,
- addition of 750 µl LB medium,
- incubation 60 min at 37 °C and 750 rpm
- plating on agar plates containing 100 µg/ml chloramphenicol
- storage over night at 37°C
Further tasks:
control cell clones
Phage Display
Investigator: Sandrina, Sabine
Time: 2011-09-14, 16:00-03:00
Aim:check if unmodified mdnA on the surface of the phages binds to an immobilized protease. interactions of microviridin L with this protease were already detected
Material/Method:
- in ELISA plate:
- coat wells with protease trypsin in NaCO3 with ph 9,4, for 4 h
- blocking in TBS with 3% BSA for 2 h
- wash 6x with TBS-T (0,005%)
- panning with phages 1:1 helper phages and phages of interest (presenting mdnA-geneIII fusion protein on their surface, 1 h)
- wash 6x with TBS-T
- elute bound phages with 0,2 M Glycin-HCl, 10 min
- neutralize with 1 M Tris (pH 7-8)
- mix eluted phages with preparatory culture of XL1-blue cells
- incubate 10 min at 37°C without shaking
- incubate 50 min at 37°C shaking
- plate on amp plates and on kan plates
Further tasks:
check cell growing
Overnight cultures for Western Blot (mdnB)
Investigators:???
| Nadine: Who did this? |
Time:2011-9-14, ???
Aim:Prepare cultures for Western Blot from pARW089, pARW071
Material:
- LB
- Amp
- glycerol stocks:
- pARW089
- pARW071
96th Labday 2011-09-15
Transformation of pSB1C3_ABC+DE and pSB1C3_D+E into XL1 blue
Investigators:Katharina
Time:7:00-8:45
Aim:Transformation of Ligation
Materials:
- competent E. coli cells XL1-Blue
- ligation products:
- pSB1C3_ABC+De + ligation control
- pSB1C3_D+E + ligation control
Method:
- addition of 2 µl ligation reaction to cells (XL1-blue) in 1.5 ml Eppi,
- incubation 20 min on ice,
- heat shock 45 sec at 42°C,
- incubation 3 min on ice,
- addition of 750 µl LB medium,
- incubation at 37 °C shaking for 60 min,
- plating on LB medium with appropriate antibiotic (Cm)
- storage @ 37°C
Further tasks:
- Picking clones for overnight culture
- Producing glycerol stocks
- miniprep
- sending for sequencing by latest Friday!!
Output:
- 4 plates with Cm:
- pSB1C3_ABC+DE
- control pSB1C3_ABC
- pSB1C3_D+E
- control pSB1C3_D
Checking fluorescence of expression backbones via fluorescence microscope
Time: 2011-09-15; 7:00-10:00
Investigators:Katharina
Materials
- liquid cultures of the expression backbones
- pSB1A3_YFP_Ara
- pSB1A3_YFP_Lac
- pSB1K3_CFP_Ara
- pSB1K3_CFP_Lac
- pSB1K3_YFP_Lac
Method
- induction using IPTG and Arabinose, respectively
- keep shaking @ 37°C for 2h
- check fluorescence of induced and uninduced, control culture with fluorescence microscope (AG Walz)
Results
- no fluorescence was detected (just autofluorescence)
Agarose Gel electrophoresis and gel extraction of mdnABC, mdnD, mdnDE, mdnD
Time: 2011-09-15
Investigators:Katharina
Materials
- digested mdn_ACD
- digested mdn_E
- digested mdn_D
- digested mdn_DE
Production of one 1% gel
- 1 % gel: 0.5 g agarose in 50 ml 1x TAE buffer
- Adding 2 µl gel red to the gel
Loading gels and running
1. 0,8 % gel
| lane | Sample | Volume in µl | Expected size in bp |
| M | Gene Ruler DNA Ladder Mix | 15µl (1:100) | |
| 1 | mdn_ABC | 30µl + 5µl 6x loading dye | |
| 2 | mdn_D | 30µl + 5µl 6x loading dye | |
| 3 | mdn_DE | 30µl + 5µl 6x loading dye | |
| 4 | mdn_E | 30µl + 5µl 6x loading dye |
- 100V
- Ca. 1h then 25V for 2h because of seminar
- samples were run out of the gel, but it was decided that we do not need these samples!
Miniprep of several overnight cultures
Investigators: Steffi, Niels, Nadja
Aim:
1. Miniprep:
- 2 overnight cultures
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- Protocol for high-copy plasmids
- elution with 50 µl elution buffer
- measuring concentration with NanoDrop:
| Sample | concentration in ng/µl | 260/280 | 260/230 |
|---|---|---|---|
| N1 | 388,8 | 1,98 | 2,27 |
| N2 | 288,3 | 1,99 | 2,16 |
| N3 | 506,9 | 2,03 | 2,45 |
| N4 | 243,0 | 1,99 | 2,37 |
| N5 | 267,2 | 1,99 | 2,43 |
| N6 | 243,4 | 2,02 | 2,32 |
| N7 | 298,9 | 1,99 | 2,20 |
| N8 | 235,4 | 1,95 | 2,08 |
| N9 | 182,2 | 1,93 | 2,68 |
| N10 | 225,5 | 1,92 | 2,34 |
| N11 | 213,8 | 1,91 | 2,41 |
| N12 | 236,9 | 1,91 | 2,19 |
| N13 | 215,1 | 1,91 | 2,32 |
| N14 | 209,2 | 1,91 | 2,32 |
| N15 | 213,0 | 1,89 | 2,01 |
| N16 | 197,6 | 1,90 | 2,07 |
- stored in -20°C (red box, expression backbones)
2. Preparation of glycerol stocks:
- adding 300 µl glycerol to 700 µl culture
further task
- sequencing
sequencing of
Investigators: Niels, Vanessa
Aim:
1. sequencing:
- 16 clones (miniprep 2011-09-15)
- 1500 ng in 20 µl:
| Sample | DNA [µl] | Water [µl] |
|---|---|---|
| N1 | 3,5 | 16,5 |
| N2 | 5 | 15 |
| N3 | 3 | 17 |
| N4 | 5,5 | 14,5 |
| N5 | 5 | 15 |
| N6 | 5,5 | 14,5 |
| N7 | 4,5 | 15,5 |
| N8 | 6 | 14 |
| N9 | 8 | 12 |
| N10 | 6 | 14 |
| N11 | 6,5 | 13,5 |
| N12 | 6 | 14 |
| N13 | 6,5 | 13,5 |
| N14 | 6,5 | 14 |
| N15 | 6 | 14 |
| N16 | 7 | 13 |
seqencing
- by GATC
Western Blot of mdnB + pARW071/pARW089
Investigators: Niels,
Aim:
detect mdnB by Immunoblot
Method
- overnight cultures spin at max speed
- supernatant discard
- cells were resuspended in 1 ml 0,5 M Tris-Hcl pH 7,4
- cells where broken by Branson Digital Sonifier
- 1s on / 3s off
| Nadine: 40 min sonification?!? that is quite long for 1 ml cell suspension... |
- 30%amplitude
- for 10 min
- spin for 30 min at 4°C by 3220xg
- 15 µl sample were mixed with 5µl 4x SDS-loading buffer
- incubated by 95°C for 5 min
- load on gradient gel
- 40V for 30min (max mA)
- 20mA until end (max V)
- gel and membrane were equilibrated for 10 minutes in Western blot transfer buffer
- blots at 200mA for 1 hour
- blocked over night in PBS-T (5% w/v milk powder) at 4°C
Materials
Western blot transfer buffer
- 15,6 mM Tris
- 120 mM Glycine
PBS-T (pH 7.4)
- 140 mM NaCl
- 2.7 mM KCL
- 8 mM Na2HPO4
- 18 mM KH2PO4
- 0,01% Tween 20
overnight culture of picked E. coli clones transformed with pSB1C3 containing mdnA
Investigators: Sabine, Sandrina
Aim: control ligation of mdnA into pSB1C3
Method/Materials:
- 10 clones from pSB1C3 with mdnA (chloramphenicol)
- 5 ml LB medium per clone
- storage over night at 37°C and 800 rpm
Further tasks:
- test digestion
97th Labday 2011-09-16
check plates - resistance of competent E. coli XL1 blue cells
Investigators: Sascha, Steffi
Aim: check resistance of competent cells - E. coli XL1 blue (from 2011-09-12, Niels, Katharina, Steffi)
Materials:
- agar plates from competent cells - E. coli XL1 blue from 2011-09-12 (Kat/Nie/Ste)
Results:
- all competent cells - E. coli XL1 blue grow on agar plates with Tetracycline
- no E. coli XL1 clones on agar plates with Ampicillin, Kanamycine, Chloramphenicol
Conclusions:
- competent cells - E. coli XL1 blue work and can be used for transformation
Antibody detection of mdnB + pARW071/pARW089
Investigators: Steffi
Aim: control if mdnB + pARW071/pARW089 is presented on the membrane
Method:
- incubate blocked membrane (Niels, 2011-09-15) for 1 h with primary antibody (anti-mdnB-antibody, 1:10000 in TBS-T)
- wash 3x 10 min with TBS-T buffer
- incubate for 1 h with secondary antibody (ZAMAK-POD, 1:5000 in TBS-T)
- wash 3x 10 min with TBS-T buffer
- develop membranes with ECL- Kit
Results:
- no signals were seen
Further tasks:
- repeat SDS-PAGE for mdnB + pARW071/pARW089
- repeat Western Blot and Antibody-Detection
Miniprep of pSB1C3+mdnB
Investigator: Katharina, Steffi
Materials:
- liquid culture of pSB1C3+mdnB
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- Protocol for high-copy plasmids
- elution with 50 µl H2O
- measuring concentration with NanoDrop:
| Sample | concentration in ng/µl |
|---|---|
| pSB1C3+mdnB 1 | |
| pSB1C3+mdnB 2 | |
| pSB1C3+mdnB 3 |
Further tasks:
- sending for sequencing
Miniprep of several overnight cultures pSB1C3+mdnA
Investigator: Katharina, Steffi
Materials:
- liquid culture of pSB1C3+mdnA
- NucleoSpin® Plasmid (NoLid) (Macherey-Nagel)
- Protocol for high-copy plasmids
- elution with 50 µl H2O
- measuring concentration with NanoDrop:
| Sample | concentration in ng/µl |
|---|---|
| pSB1C3+mdnA 1 | 323.7 |
| pSB1C3+mdna 2 | 309.3 |
| pSB1C3+mdna 3 | 342.8 |
| pSB1C3+mdnA 4 | 326.7 |
| pSB1C3+mdnA 5 | 248.8 |
| pSB1C3+mdnA 6 | 261.8 |
| pSB1C3+mdnA 7 | 266.6 |
| pSB1C3+mdnA 8 | 267.7 |
| pSB1C3+mdnA 9 | 229.7 |
| pSB1C3+mdnA 10 | 305.1 |
Further tasks:
- test digestion for confirmation
- sending for sequencing
Send clones 4,5,8,10 (mdnA in pSB1C3) for sequencing (after test digestion)
Investigators: Sabine, Sandrina
Aim:control ligation of mdnA and geneIII into pSB1C3 for generating biobricks
Method/Materials:
- Primer: VF2
- 20 µl with 70 ng/µl DNA
Further tasks:
check alignment
Repeated Phage Display with another trypsin protease
Investigator: Sandrina, Sabine
Time: 2011-09-16, 9:00-17:00
Aim:check if unmodified mdnA on the surface of the phages binds to an immobilized protease. interactions of microviridin L with this protease were already detected
Material/Method:
- in ELISA plate:
- coat wells with protease trypsin in NaCO3 with ph 9,4, for 4 h
- blocking in TBS with 3% BSA for 2 h
- wash 6x with TBS-T (0,005%)
- panning with phages 1:1 helper phages and phages of interest (presenting mdnA-geneIII fusion protein on their surface, 1 h)
- wash 6x with TBS-T
- elute bound phages with 0,2 M Glycin-HCl, 10 min
- neutralize with 1 M Tris (pH 7-8)
- mix eluted phages with preparatory culture of XL1-blue cells
- store over night at 4°C
Further tasks:
check binding of phages by plating them
Digest of pSB1A3 + YFP clone III, ligation with promoters and transformation
Investigators: Nadine, Jessica, Niels
Time: 2011-09-16, 16:00-22:00
Material:
- pSB1A3_YFP clone III, 15.08.11, Steffi
- digested promoters: Ara 1 (NW, 16.08.11), lac 1(NW, 16.08.11), Ara 2, lac 2
Digest:
- 30 µl reaction:
- 10 µl pSB1A3_YFP
- 3 µl NEB Buffer 4
- 0.3 µl BSA
- 1.5 µl EcoRI-HF
- 1.5 µl XbaI
- 13.7 µl water
- 37°C for 1.5 h
Gel electrophoresis:
| lane | Sample | Volume in µl | Expected size in bp |
|---|---|---|---|
| M | GeneRuler™ DNA Ladder Mix (diluted 1:10) | 20 | |
| 1 | - | - | |
| 2 | digest of pSB1A3+YFP | 36 |
[[File:|200px]]
Gel extraction:
- using NucleoSpin Extact II
- elution with 50 µl NE buffer
Ligation:
- 1 µl T4 ligase buffer
- 1 µl T4 ligase
- 4 µl backbone
- 4 µl insert
Transformation:
| Nadine: please do not forget to write about the transformation protocol (I think it was Niels´ task?!?) |
Output:
- amp-LB agar plates
- pSB1A3_YFP_Ara1
- pSB1A3_YFP_Ara2
- pSB1A3_YFP_lac1
- pSB1A3_YFP_lac2
- pSB1A3_YFP control
Further task:
- picking clones for overnight plates
98th Labday 2011-09-17
Miniprep of pSB1C3+mdn_ABC+DE and pSB1C3+mdnABC
Investigator: Katharina
Materials:
- liquid culture of pSB1C3+mdn_ABC+DE and pSB1C3+mdnABC
- Buffer A1, A2, A3 of the Macherey-Nagel Kit
- Isopropanol
- 70% EtOH
Method:
- pellet the cells
- resuspend in 150 µl Buffer A1
- add 150 µl Buffer A2 and invert the tube
- incubate @ room temperature for 5 min
- add 150 µl Buffer A3 and invert the tube
- centrifuge for 5 min @ 11000 rpm
- transfer supernatant to a new tube
- add 1000 µl isopropanol
- vortex and keep at room temperature for 5 min
- centrifuge for 5-10 min @ 11000 rpm
- discard the supernatant and wash the pellet in 70% ethanol
- centrifuge for 5-10 min @ 11000 rpm
- discard the supernatant
- dry the pellet (either @ room temperature or @ 37°C)
- resuspend the pellet in 30 µl sterile water
- measuring concentration with NanoDrop:
| Sample | concentration in ng/µl | 260/280 | 260/230 |
|---|---|---|---|
| mdnABC 1 | 4013.3 | 2.07 | 2.19 |
| mdnABC 2 | 4549.2 | 2.06 | 2.21 |
| mdn_ABC+DE 1 | 3691.1 | 2.03 | 2.20 |
| mdn_ABC+DE | 3012.9 | 1.97 | 2.07 |
- stored in green mdn-biobrick box (eppis have yellow labels on top)
Checking plates from yesterday (Expression Backones)and picking clones
Investigator: Nadine
Time: 2011-9-17, ~15:00
Materials:
- amp-LB agar plates (2011-9-16, Niels)
- pSB1A3_YFP_Ara1
- pSB1A3_YFP_Ara2
- pSB1A3_YFP_lac1
- pSB1A3_YFP_lac2
- pSB1A3_YFP control
- LB
- ampicillin
Results:
- control plate: ~40 clones
- ligation plates: ~ 160 clones
Protocol:
- 5 ml LB
- add 5 µl Amp
- pick 3 colonies per plate and add to LB medium
Output:
- 12 overnight cultures:
- pSB1A3_YFP_Ara2, Klon 1, 2 and 3
- pSB1A3_YFP_Ara1, Klon 4, 5 and 6
- pSB1A3_YFP_lac2, Klon 7, 8 and 9
- pSB1A3_YFP_lac2, Klon 10, 11 and 12
Further tasks:
- miniprep tomorrow ( DO NOT FORGET THE GLYCEROL STOCKS)
- overnight cultures for monday for fluorescence tests
- sequencing on monday?!?!
Overnight cultures for Western Blot
Investigator: Nadine
Time: 2011-9-17, ~15:00
Materials:
glycerol stocks:
- pARW089
- pARW071
- pUP089 (#G2)
- pSB1C3+mdnABCDE (#G61 and #G62)
- LB
- ampicillin
- chloramphenicol
- kanamycin
Protocol:
- 10 ml LB
- add 10 µl antiobiotics
- add cells
- incubate overnight at 37°C and 200 rpm
Control of repeated Phage Display with another trypsin protease
Investigator: Sandrina, Sabine
Time: 2011-09-17, 11:00-18:00
Aim:check if unmodified mdnA on the surface of the phages binds to an immobilized protease. interactions of microviridin L with this protease were already detected
Material/Method:
- over night culture of XL1-blue cells was infected with eluted phages
- 1000 cells were infected with 10.000 phages and plated on agar with kanamycin and on agar with ampicillin
- control: 1:1 mix of helperphages and produced phages of interest carrying mdnA-geneIII plated also on agar with kanamycin and on agar with ampicillin
Further tasks:
- check cell growth
Miniprep of 14_3C2,3,4 and TEv 2,3,4
For better understanding of described experiment see also: http://141.89.201.101/iGEM/wiki2011/images/c/c8/UP_SG_Klonierungsschema_Protease.pptx
Investigators: Sascha, Stefan, Sebastian
Aim:
get plasmids
Methode:
- pellet the cells
- resuspend in 150 µl Buffer A1
- add 150 µl Buffer A2 and invert the tube
- incubate @ room temperature for 5 min
- add 150 µl Buffer A3 and invert the tube
- centrifuge for 5 min @ 11000 rpm
- transfer supernatant to a new tube
- add 1000 µl isopropanol
- vortex and keep at room temperature for 5 min
- centrifuge for 5-10 min @ 11000 rpm
- discard the supernatant and wash the pellet in 70% ethanol
- centrifuge for 5-10 min @ 11000 rpm
- discard the supernatant
- dry the pellet (either @ room temperature or @ 37°C)
- resuspend the pellet in 30 µl sterile water
Further Tasks:
send plasmids for sequenzing
99th Labday 2011-09-18
Miniprep of pSB1A3_YFP Ara und Lac cultures
Time: 2011-09-18
Investigator: Nadja
Materials:
- liquid culture: * 12 overnight cultures:
- pSB1A3_YFP_Ara2, Klon 1, 2 and 3
- pSB1A3_YFP_Ara1, Klon 4, 5 and 6
- pSB1A3_YFP_lac2, Klon 7, 8 and 9
- pSB1A3_YFP_lac2, Klon 10, 11 and 12
- Buffer A1, A2, A3 of the Macherey-Nagel Kit
- Isopropanol
- 70% EtOH
Method:
- pellet the cells
- resuspend in 150 µl Buffer A1
- add 150 µl Buffer A2 and invert the tube
- incubate @ room temperature for 5 min
- add 150 µl Buffer A3 and invert the tube
- centrifuge for 5 min @ 11000 rpm
- transfer supernatant to a new tube
- add 1000 µl isopropanol
- vortex and keep at room temperature for 5 min
- centrifuge for 5-10 min @ 11000 rpm
- discard the supernatant and wash the pellet in 500µl 70% ethanol
- centrifuge for 5-10 min @ 11000 rpm
- discard the supernatant
- dry the pellet (either @ room temperature or @ 37°C)
- resuspend the pellet in 30 µl sterile water
- measuring concentration with NanoDrop:
| Sample | concentration in ng/µl | 260/280 | 260/230 |
|---|---|---|---|
| pSB1A3_YFP_Ara2 Clone1 | 718, 8 | 1,9 | 1,74 |
| pSB1A3_YFP_Ara2 Clone2 | 5360, 5 | 1,9 | 1,95 |
| pSB1A3_YFP_Ara2 Clone3 | 5882, 9 | 2,0 | 1,86 |
| pSB1A3_YFP_Ara1 Clone4 | 668 | 1, 2 | 1,14 |
| pSB1A3_YFP_Ara1 Clone5 | 5570, 3 | 1,9 | 1,58 |
| pSB1A3_YFP_Ara1 Clone6 | 7170,3 | 2,0 | 1,93 |
| pSB1A3_YFP_Lac2 Clone7 | 3275,8 | 5,7 | -3.41 |
| pSB1A3_YFP_Lac2 Clone8 | 4471,6 | 2,0 | 1,61 |
| pSB1A3_YFP_Lac2 Clone9 | 4577,7 | 2,1 | 1,97 |
| pSB1A3_YFP_Lac1 Clone10 | 3535,8 | 1,9 | 1,5 |
| pSB1A3_YFP_Lac1 Clone11 | 3904,6 | 2,1 | 2,22 |
| pSB1A3_YFP_Lac1 Clone12 | 5395,4 | 2,0 | 1,91 |
- stored in a new green expression backbones box at -20°C
Further tasks:
- overnight cultures for fluorescence measurement
- sending for sequencing
Western Blot of mdnB + pARW071/pARW089
Time: 2011-09-18
Investigator: Sebastian, Nadja
Method:
- overnight cultures spin at max speed
- supernatant discard
- cells were resuspended in 1 ml 0,5 M Tris-Hcl pH 7,4
- cells where broken by Branson Digital Sonifier
- 2s on / 2s off
- 30%amplitude
- for 2min
- spin for 10 min by 17.0 xg
- 15 µl sample were mixed with 5µl 4x SDS-loading buffer
- incubated by 95°C for 5 min
- load on gradient gel
- 50V for 30min (max mA)
- 40mA until end (max V)
- one gel and membrane were equilibrated for 10 minutes in Western blot transfer buffer
- membrane was immobilized with methanol
- blots at 200mA for 1 hour
- blocked over night in PBS-T (5% w/v milk powder) at 4°C
- second gel was stained and shaked over night in Coomassie
Materials:
1. Western blot transfer buffer
- 15,6 mM Tris
- 120 mM Glycine
2. PBS-T (pH 7.4)
- 140 mM NaCl
- 2.7 mM KCL
- 8 mM Na2HPO4
- 18 mM KH2PO4
- 0,01% Tween 20
Further tasks:
- washing steps and treatment with first and second antibody for immunodetection of mdnB
Antibody detection of mdnB + pARW071/pARW089
Investigators: Steffi
Aim: control if mdnB + pARW071/pARW089 is presented on the membrane
Method:
- incubate blocked membrane (Nadja, Sebastian, 2011-09-16) for 1 h with primary antibody (anti-mdnB-antibody, 1:10000 in TBS-T)
- wash 3x 10 min with TBS-T buffer
- incubate for 1 h with secondary antibody (ZAMAK-POD, 1:5000 in TBS-T)
- wash 3x 10 min with TBS-T buffer
- develop membranes with ECL- Kit
Results:
- no signals were seen
Further tasks:
- repeat SDS-PAGE for mdnB + pARW071/pARW089
- repeat Western Blot and Antibody-Detection
99th Labday 2011-09-20
Miniprep of pSB1A3_YFP Ara-cultures
Investigator: Steffi
Materials:
- 2 overnight cultures
- pSB1A3_YFP Ara1 I
- pSB1A3_YFP Ara2 II
- Buffer A1, A2, A3 of the Macherey-Nagel Kit
- Isopropanol
- 70% EtOH
Method:
- pellet the cells
- resuspend in 250 µl Buffer A1
- add 250 µl Buffer A2 and invert the tube
- incubate @ room temperature for 5 min
- add 250 µl Buffer A3 and invert the tube immediately
- centrifuge for 10 min @ 11000 rpm
- transfer supernatant to a new tube
- add 0,7 volume of isopropanol
- invert the tupe and keep at room temperature for 5 min
- centrifuge for 10 min @ 11000 rpm
- discard the supernatant and wash the pellet in 500µl 70% ethanol
- centrifuge for 10 min @ 11000 rpm
- discard the supernatant
- dry the pellet (either @ room temperature or @ 37°C)
- resuspend the pellet in 30 µl sterile water
- measuring concentration with NanoDrop:
| Sample | concentration in ng/µl | 260/280 | 260/230 |
|---|---|---|---|
| pSB1A3_YFP Ara1 I | 3441,1 | 2,0 | 2,26 |
| pSB1A3_YFP Ara2 II | 4874,4 | 2,11 | 2,39 |
- stored in a new green expression backbones box at -20°C
Further tasks:
- overnight cultures for fluorescence measurement
- sending for sequencing
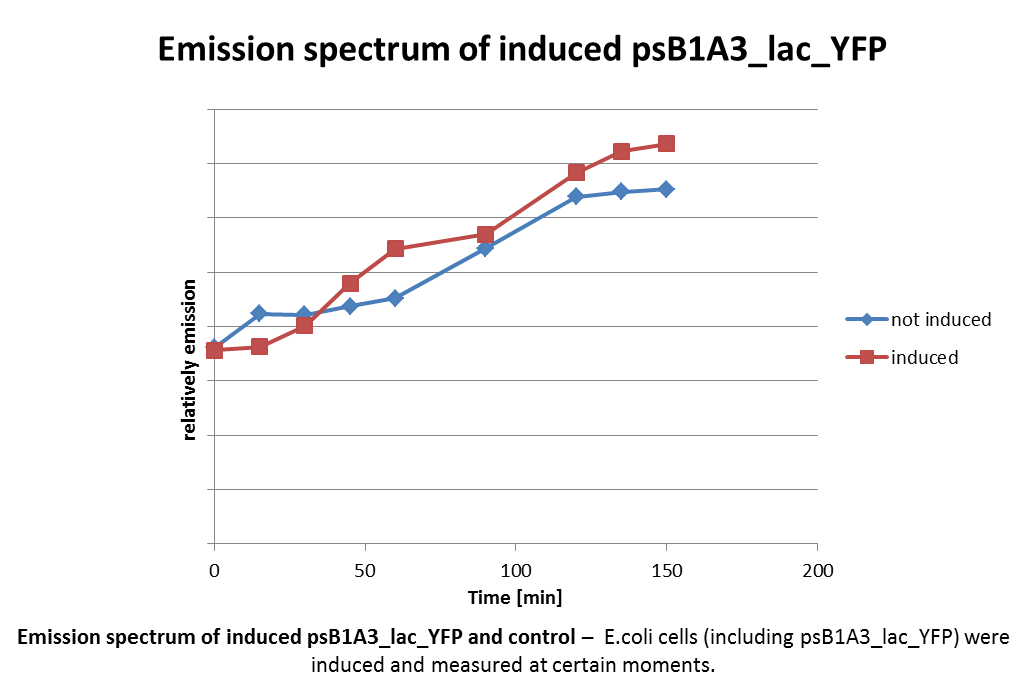
Analysis of psB1A3_Ava_YFP and psB1A3_IPTG_YFP by fluorescence spectroscopy.
Investigators: Niels, Jessica
Materials:
- psB1A3_Ava_YFP over night culture
- psB1A3_IPTG_YFP over night culture
Methods:
- 1 ml culture were transfered in new media
- observed growing by measuring OD(600nm)
- inducing of promotor by 5 mM Ara or 1 mM IPTG at a OD(600nm) by 0.4-0.5)
- measuring of samples every 15 mintes by fluorescence spectroscopy
further tasks:
- analyze by fluorescence microscopy
Analysis of psB1A3_Ava_YFP and psB1A3_IPTG_YFP by fluorescence microscopy.
Investigators: Katharina
Materials:
- psB1A3_Ava_YFP over night culture
- psB1A3_IPTG_YFP over night culture
Methods:
- 1 ml culture were transfered in new media
- observed growing by measuring OD(600nm)
- inducing of promotor by 5 mM Ara or 1 mM IPTG at a OD(600nm) by 0.4-0.5)
- measuring of samples by fluorescence microscopy (1h after inducing)
Results:
Sending of BioBricks to MIT
Investigators: Nicole, Nadine, Sebastian
BioBricks
| Label number | BioBrick Nickname | Tube label | c [ng/µl] |
|---|---|---|---|
| BBa_K627000 | mdnED | 1 | 25 |
| BBa_K627001 | mdnA | 2 | 25 |
| BBa_K627002 | mdnB | 3 | 25 |
| BBa_K627003 | mdnC | 4 | 25 |
| BBa_K627004 | mdnD | 5 | 25 |
| BBa_K627005 | mdnE | 6 | 25 |
| BBa_K627006 | mdnA c-myc gene III | 7 | 25 |
| BBa_K627007 | c-myc gene III | 8 | 25 |
| BBa_K627008 | AraC TEV protease 1 | 9 | 25 |
| BBa_K627009 | AraC TEV protease 2 | 10 | 25 |
| BBa_K627010 | AraC TEV protease 3 | 11 | 25 |
| BBa_K627011 | AraC 14_3C protease | 12 | 25 |
| BBa_K627012 | ssTorA CS-TEV BlaFL | 13 | 25 |
| BBa_K627013 | ssTorA CS-14_3C BlaFL | 14 | 25 |
| BBa_K627014 | A3_Ara_YFP clone 1 | 15 | 25 |
| BBa_K627015 | A3_lac_YFP clone 2 | 16 | 25 |
 "
"