Team:Potsdam Bioware/Project/Summary
From 2011.igem.org
(→Highlights) |
(→Microviridin) |
||
| (112 intermediate revisions not shown) | |||
| Line 4: | Line 4: | ||
===Modification, Selection and Production of Cyclic Peptides for Therapy=== | ===Modification, Selection and Production of Cyclic Peptides for Therapy=== | ||
| - | One key task of biopharmaceuticals is the binding and blocking of deregulated proteins. | + | One key task of biopharmaceuticals is the binding and blocking of deregulated proteins. Picking the right lead structure for biopharmaceuticals is very important for success. This year, we developed a novel system for the modification, selection and optimization of peptides showing potential for protease inhibition. These promising inhibitors are found in cyanobacteria and are called Microviridins. They belong to a class of peptides characterized by unusual ω-ester and ω-amide bonds between the amino-acid side chains, which are also referred to as depsipeptides. These modifications are introduced post translationally by a set of enzymes and result in an extraordinary tricyclic cage structure. |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<br> | <br> | ||
| - | |||
| - | |||
<br> | <br> | ||
| - | + | Proteases are a large enzyme family and comprise about 647 human gene products. Therefore they are important drug targets. In addition, many harmful bacteria, viruses and fungi use proteases in their reproduction cycle and growth. The ability to block these proteases is highly relevant for therapy. Prominent examples of targets are the angiotensin-converting enzyme (ACE) and HIV proteases. | |
| + | <br><br>We chose a 6.5 kb Microviridin gene cluster named ''mdnABCDE'' from ''Mycrocystis aeruginosa'' NIES843. The <i>mdn</i> gene cluster comprises (i) a gene encoding a precursor peptide named MdnA, which is then modified to form the Microviridin, (ii) two genes encoding ATP-grasp-type ligases named ''mdnB'' and ''mdnC'', (iii) an ABC- transporter encoding gene named ''mdnE'' as well as (iv) one gene encoding an N- acetyltransferase of the GNAT family named ''mdnD'' (Ziemert et al., 2008). Our major aim was to modify the MdnA precursor peptide and optimize its protease inhibiting properties. Towards this goal, we synthesized semi-rational ''mdnA'' gene libraries with partially randomized oligonucleotides. One library was cloned, verified by sequencing, and successfully used for selection. | ||
<br> | <br> | ||
| - | |||
<br> | <br> | ||
| - | + | To identify the best candidates inhibiting various given proteases, we established two different selection systems: Phage Display and a new developed ''in-vivo'' selection assay. Phage display is a frequently used technique in laboratories, yet to our knowledge, phage display of cellularly cyclized peptides is new. We constructed a fusion between the surface protein geneIII of the phage and the ''mdnA'' gene within the ''mdnA'' gene cluster. This system was verified by western blotting, Phage-ELISA and controlled phage panning experiments. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
<br> | <br> | ||
| - | |||
<br> | <br> | ||
| - | + | We also devised and constructed a recombinant ''in-vivo'' selection system linking protease degradation to antibiotic resistance. This system is divided into three parts: first a protease activity detector device, second a protease generator device, and third a protease blocking device. For the protease activity detector device we fused various protease cleavage sites between a signal sequence and the antibiotic resistance conferring enzyme β-lactamase. β-lactamase provides resistance when its transferred into the periplasm. Thus, when the protease cleaves the signal sequence from the β-lactamase antibiotic resistance is abolished and cells die under selective pressure. However, when our Microviridin variants inhibit the protease, cells survive, allowing for an easy and efficient selection of protease inhibitors. We demonstrated a wide dynamic range of the system between expressed and non-expressed protease in the presence of increasing ampicillin concentrations. Importantly, after co-transformation with our ''mdnA''-libary we were able to isolate several clones, which are currently being characterized. | |
<br> | <br> | ||
| - | |||
<br> | <br> | ||
| - | In addition to the practical work we | + | In addition to the practical work, we established a mathematical model of the ''in-vivo'' selection system. This model, which we were able to fit to experimental data, helped us to understand the selection process. We modeled the reaction kinetics with ordinary differential equations and simulated and fitted data with matlab. |
| - | + | <br> | |
| - | + | <br> | |
| - | + | Last but not least, we had several human practice projects. We send a survey to all members of the German parliament, visited a member of the parliament, held seminars on ethics and invited children to the lab. | |
| + | <br> | ||
| + | *Ziemert, N., Ishida, K., Liaimer, A., Hertweck, C. & Dittmann, E. (2008). Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angewandte Chemie (International ed. in English) 47, 7756-9 | ||
| + | <br><br> | ||
== Highlights == | == Highlights == | ||
| Line 39: | Line 29: | ||
=== Microviridin === | === Microviridin === | ||
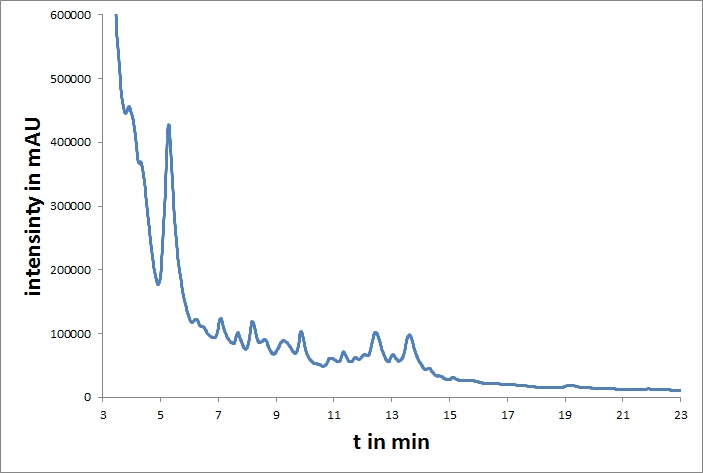
| - | [[File: | + | [[File:UP_HPLC_mvd_01.jpg|left|300px|thumb|'''Figure 1:''' <b>HPLC chromatogram of heterologously expressed mdnA</b>]] |
| - | The | + | <br> |
| - | + | The microviridin sub-project aimed for modifications at the genetic level such that protease inhibiting activities of the resulting peptides are enhanced. We used random mutagenesis or focused randomization of oligonucleotides for the creation of gene libraries, which can be screened for <i>mdnA</i>-variants with therapeutically promising mutations. For further experiments, we also fused mdnA to a myc-tag.<br> | |
| - | + | In addition, all coding genes of the <i>mdn</i>-cluster were converted to BioBricks. The microviridin from <i>mdnA</i> was purified and characterized by HPLC and cyclization was verified by mass spectrometry.<br> | |
| + | For the quick expression of several library clones, we also built auxiliary plasmid backbones with inducible promoters according to a proposed extension of the iGEM cloning standard. | ||
| + | '''[[:Team:Potsdam_Bioware/Project/Details_Microviridin|[more]]]''' | ||
| + | <br> | ||
| + | <br> | ||
<br> | <br> | ||
=== Phage Display === | === Phage Display === | ||
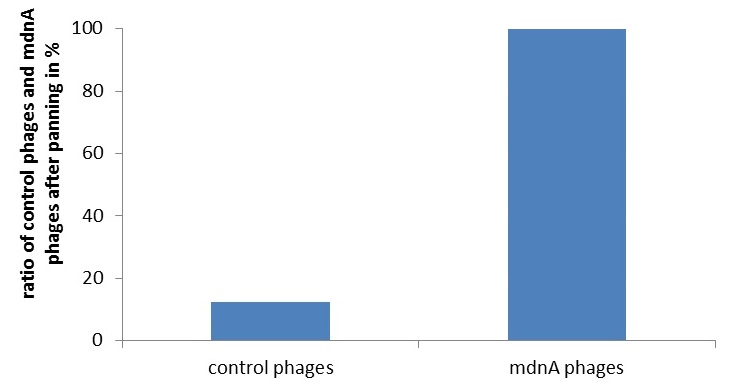
| - | + | [[File:UP_Phage_Display_02.jpg|left|290px|thumb|'''Figure 2: Phage Display of microviridin.''' Enrichment of MdnA carrying phages upon panning towards a protease was demonstrated.]] | |
| - | Phage | + | <br> |
| + | Phage display is a powerful tool for selecting peptides or proteins that bind and regulate the function of target proteins. Phages robustly link the genotype to the phenotype of the presented peptide allowing for selection of protease binders under controlled ''in vitro'' conditions. We established genetics and panning of therapeutically interesting ''mdnA''-libraries towards a panel of several purified proteases. First, the general suitability of phage display for this purpose was shown. An appropriate phagemid vector harboring an <i>mdnA-myc-geneIII</i> fusion gene, was constructed. The expression of the MdnA-myc-geneIII protein in ''E. coli'' was shown by western blot. Production of phage particles carrying MdnA were analyzed by Phage-ELISA. Finally, phage display procedures were optimized and an enrichment of MdnA modified phages over a control was shown. '''[[:Team:Potsdam_Bioware/Project/Details_Phage|[more]]]''' | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
=== In Vivo Selection === | === In Vivo Selection === | ||
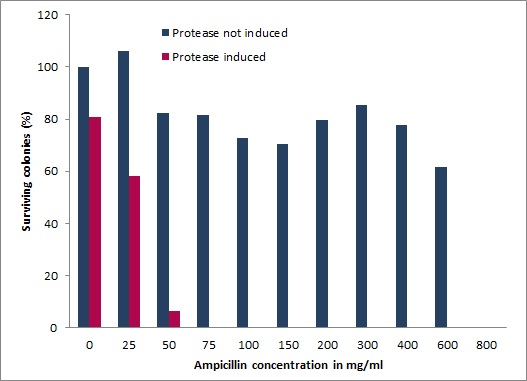
| + | [[Image:UP_SurvTEV_onepla.jpg|left|290px|thumb|'''Figure 3: Ampicillin resistance test.''' | ||
| + | Survival screen, without induced (blue) and with induced (magenta) protease. From left to right increasing ampicillin concentrations are shown.]] | ||
| - | + | A proper designed ''in vivo'' selection is very powerful, quick and entails an inherent selection against general toxicity. We designed and employed a multi-component system enabling inexpensive and time-saving selection of microviridin libraries blocking various proteases. Our genetic circuit links with high sensitivity and wide dynamic range a protease generator, a protease detector and a protease inhibitor device. This is achieved by coupling the export of the enzyme ß-lactamase to a signal sequence via an interspersed modularly cloned protease cleavage site. Inducible lactamase and protease generators are united on a single plasmid while the <i>mdnA</i> library is provided in trans. Functionality of the system was characterized under a multitude of conditions for different proteases. Ultimately, a one step selection of a genetic <i>mdnA</i> library against the protease TEV with increasing concentrations of the antibiotic ampicillin yielded inhibiting clones. '''[[:Team:Potsdam_Bioware/Project/Details_Selection|[more]]]''' | |
| - | + | <br><br> | |
| - | + | <br><br> | |
| + | <br><br><br> <br> | ||
| - | + | === Modeling === | |
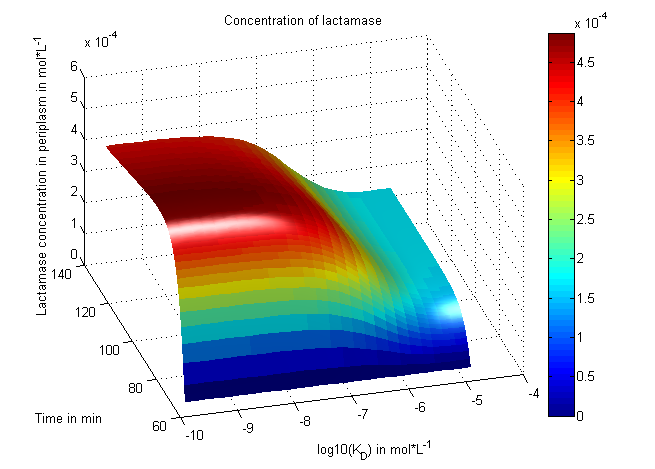
| - | + | [[File: UP_3Dplot_Lact_06.png|left|300px|thumb|'''Figure 4:''' <b>Plot predicting lactamase concentrations in the periplasm</b> (and thus the cell fittness) in dependence of the enzyme inhibition reaction coefficient K<sub>D</sub>.]] | |
| + | <br> | ||
| + | There is no synthetic biology without modeling, of course. We focused on systems modeling of our ''in vivo'' selection system in which reaction kinetics are analyzed and outcomes are predicted or fitted to experimental data. On one hand, predictions based on known parameters enable the rational choice of optimized conditions (e.g. induction levels of the protease generator). On the other hand, fitting to biological readouts (e.g. survival) enables deciphering of intrinsic parameters (e.g. interactions constants).<br><br> | ||
| + | Reactions were coded as ordinary differential equations under consideration of sequential induction time points. Substance concentrations were numerically propagated through time. A useful dynamic range and also a certain robustness of our system were confirmed. We learned about correct time-scales for triggering and we predicted cell-division rates as a reference for the lab work. In a final step, we fitted our model to wet-lab measurements. '''[[:Team:Potsdam_Bioware/Project/Details_Modeling|[more]]]'''<br><br><br><br><br> | ||
| + | === Ethics === | ||
| - | + | For information about an ethics seminar and a survey among politicians see '''[[:Team:Potsdam_Bioware/Safety_Ethics|[here]]]'''. | |
| - | + | ||
| - | + | === Software === | |
| - | + | ||
| - | === | + | |
| - | + | Have a look at the features and screenshots of our BioLog app on this '''[[:Team:Potsdam_Bioware/Software|[page]]]'''. | |
| - | + | ||
Latest revision as of 03:02, 29 October 2011
Summary
Modification, Selection and Production of Cyclic Peptides for Therapy
One key task of biopharmaceuticals is the binding and blocking of deregulated proteins. Picking the right lead structure for biopharmaceuticals is very important for success. This year, we developed a novel system for the modification, selection and optimization of peptides showing potential for protease inhibition. These promising inhibitors are found in cyanobacteria and are called Microviridins. They belong to a class of peptides characterized by unusual ω-ester and ω-amide bonds between the amino-acid side chains, which are also referred to as depsipeptides. These modifications are introduced post translationally by a set of enzymes and result in an extraordinary tricyclic cage structure.
Proteases are a large enzyme family and comprise about 647 human gene products. Therefore they are important drug targets. In addition, many harmful bacteria, viruses and fungi use proteases in their reproduction cycle and growth. The ability to block these proteases is highly relevant for therapy. Prominent examples of targets are the angiotensin-converting enzyme (ACE) and HIV proteases.
We chose a 6.5 kb Microviridin gene cluster named mdnABCDE from Mycrocystis aeruginosa NIES843. The mdn gene cluster comprises (i) a gene encoding a precursor peptide named MdnA, which is then modified to form the Microviridin, (ii) two genes encoding ATP-grasp-type ligases named mdnB and mdnC, (iii) an ABC- transporter encoding gene named mdnE as well as (iv) one gene encoding an N- acetyltransferase of the GNAT family named mdnD (Ziemert et al., 2008). Our major aim was to modify the MdnA precursor peptide and optimize its protease inhibiting properties. Towards this goal, we synthesized semi-rational mdnA gene libraries with partially randomized oligonucleotides. One library was cloned, verified by sequencing, and successfully used for selection.
To identify the best candidates inhibiting various given proteases, we established two different selection systems: Phage Display and a new developed in-vivo selection assay. Phage display is a frequently used technique in laboratories, yet to our knowledge, phage display of cellularly cyclized peptides is new. We constructed a fusion between the surface protein geneIII of the phage and the mdnA gene within the mdnA gene cluster. This system was verified by western blotting, Phage-ELISA and controlled phage panning experiments.
We also devised and constructed a recombinant in-vivo selection system linking protease degradation to antibiotic resistance. This system is divided into three parts: first a protease activity detector device, second a protease generator device, and third a protease blocking device. For the protease activity detector device we fused various protease cleavage sites between a signal sequence and the antibiotic resistance conferring enzyme β-lactamase. β-lactamase provides resistance when its transferred into the periplasm. Thus, when the protease cleaves the signal sequence from the β-lactamase antibiotic resistance is abolished and cells die under selective pressure. However, when our Microviridin variants inhibit the protease, cells survive, allowing for an easy and efficient selection of protease inhibitors. We demonstrated a wide dynamic range of the system between expressed and non-expressed protease in the presence of increasing ampicillin concentrations. Importantly, after co-transformation with our mdnA-libary we were able to isolate several clones, which are currently being characterized.
In addition to the practical work, we established a mathematical model of the in-vivo selection system. This model, which we were able to fit to experimental data, helped us to understand the selection process. We modeled the reaction kinetics with ordinary differential equations and simulated and fitted data with matlab.
Last but not least, we had several human practice projects. We send a survey to all members of the German parliament, visited a member of the parliament, held seminars on ethics and invited children to the lab.
- Ziemert, N., Ishida, K., Liaimer, A., Hertweck, C. & Dittmann, E. (2008). Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angewandte Chemie (International ed. in English) 47, 7756-9
Highlights
Microviridin
The microviridin sub-project aimed for modifications at the genetic level such that protease inhibiting activities of the resulting peptides are enhanced. We used random mutagenesis or focused randomization of oligonucleotides for the creation of gene libraries, which can be screened for mdnA-variants with therapeutically promising mutations. For further experiments, we also fused mdnA to a myc-tag.
In addition, all coding genes of the mdn-cluster were converted to BioBricks. The microviridin from mdnA was purified and characterized by HPLC and cyclization was verified by mass spectrometry.
For the quick expression of several library clones, we also built auxiliary plasmid backbones with inducible promoters according to a proposed extension of the iGEM cloning standard.
[more]
Phage Display
Phage display is a powerful tool for selecting peptides or proteins that bind and regulate the function of target proteins. Phages robustly link the genotype to the phenotype of the presented peptide allowing for selection of protease binders under controlled in vitro conditions. We established genetics and panning of therapeutically interesting mdnA-libraries towards a panel of several purified proteases. First, the general suitability of phage display for this purpose was shown. An appropriate phagemid vector harboring an mdnA-myc-geneIII fusion gene, was constructed. The expression of the MdnA-myc-geneIII protein in E. coli was shown by western blot. Production of phage particles carrying MdnA were analyzed by Phage-ELISA. Finally, phage display procedures were optimized and an enrichment of MdnA modified phages over a control was shown. [more]
In Vivo Selection
A proper designed in vivo selection is very powerful, quick and entails an inherent selection against general toxicity. We designed and employed a multi-component system enabling inexpensive and time-saving selection of microviridin libraries blocking various proteases. Our genetic circuit links with high sensitivity and wide dynamic range a protease generator, a protease detector and a protease inhibitor device. This is achieved by coupling the export of the enzyme ß-lactamase to a signal sequence via an interspersed modularly cloned protease cleavage site. Inducible lactamase and protease generators are united on a single plasmid while the mdnA library is provided in trans. Functionality of the system was characterized under a multitude of conditions for different proteases. Ultimately, a one step selection of a genetic mdnA library against the protease TEV with increasing concentrations of the antibiotic ampicillin yielded inhibiting clones. [more]
Modeling
There is no synthetic biology without modeling, of course. We focused on systems modeling of our in vivo selection system in which reaction kinetics are analyzed and outcomes are predicted or fitted to experimental data. On one hand, predictions based on known parameters enable the rational choice of optimized conditions (e.g. induction levels of the protease generator). On the other hand, fitting to biological readouts (e.g. survival) enables deciphering of intrinsic parameters (e.g. interactions constants).
Reactions were coded as ordinary differential equations under consideration of sequential induction time points. Substance concentrations were numerically propagated through time. A useful dynamic range and also a certain robustness of our system were confirmed. We learned about correct time-scales for triggering and we predicted cell-division rates as a reference for the lab work. In a final step, we fitted our model to wet-lab measurements. [more]
Ethics
For information about an ethics seminar and a survey among politicians see [here].
Software
Have a look at the features and screenshots of our BioLog app on this [page].
 "
"