Team:Potsdam Bioware/Project/Details Phage
From 2011.igem.org
(→References) |
|||
| (70 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{:Team:Potsdam_Bioware/Head}}{{:Team:Potsdam_Bioware/jquery}}{{:Team:Potsdam_Bioware/menu_home}} | {{:Team:Potsdam_Bioware/Head}}{{:Team:Potsdam_Bioware/jquery}}{{:Team:Potsdam_Bioware/menu_home}} | ||
| - | + | == Phage Display == | |
| - | < | + | ===Introduction=== |
| - | < | + | |
| - | + | Phage Display is an efficient tool for selecting proteins or peptides with specific binding properties from a large recombinant library. In phage display proteins are presented on the surface of bacteriophages. This enables the coupling of phenotype and stable packaged genotype, because the proteins which form the phage including the proteins of interest are coded in its genome. To test the suitability of phage display system as an appropriate screening method for recombinant <i>mdnA</i> libraries a vector containing a <i>mdnA-myc-geneIII</i>-fusion gene was generated. This vector contains a plasmid origin of replication, so it can be amplified like plasmids. Additionally it contains a f1 ori which enables the packaging of single strand DNA into phages. The vector also contains the whole <i>mdn</i>-cluster which is needed to produce the MdnA peptide. Between <i>mdnA</i> and <i>gene III</i> is located a <i>myc</i>-tag, which is used for an easy detection. The successful expression of the MdnA-myc-geneIII-fusion protein on the surface of the phage was determined by ELISA using anti-myc antibody 9E10 after transforming ''E. coli'' cells and purifying the produced phages. The next step was to perform a phage display on a known target of the MdnA. To test the fundamental suitability of this screening method, phages presenting MdnA on their surface and phages not presenting MdnA in a ratio of one to one were incubated with immobilized trypsin which is known as a target of MdnA. After the first panning round a notable concentrating of phages carrying MdnA was recognized.<br> | |
| - | The phage display vector pPDV089 was derived from the plasmid pARW089 which carries the whole | + | |
| - | First | + | ===Cloning strategy=== |
| + | |||
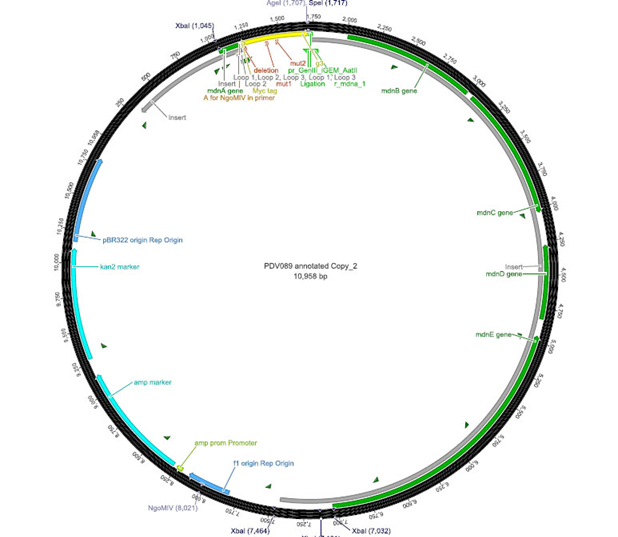
| + | The phage display vector pPDV089 was derived from the plasmid pARW089 which carries the whole <i>mdn</i>-cluster. This plasmid contains a plasmid origin of replication and additionally a f1 ori which enables the packaging of single strand DNA into phages. For selective amplification ampcillin and kanamycin resistance genes are included. To create the phagemid pPDV089 standard cloning procedure were performed. | ||
| + | First <i>mdnA</i> was excised using the restriction enzymes <i>Sfo</i>I and <i>Aat</i>II. The next step comprised the introduction of a <i>mdnA-geneIII</i>-fusion gene. Therefore <i>gene III</i> was amplified from pak100blaKDIR and <i>mdnA</i> from pARW089 by PCR. The primers were designed to enable the introduction of iGEM and other restriction sites required for further cloning steps. The purified PCR product <i>geneIII</i> was digested by <i>NgoM</i>IV and <i>Aat</i>II whereas the PCR product <i>mdnA</i> was digested by <i>Sfo</i>I and <i>Age</i>I. Subsequent the three fragment ligation of <i>mdnA</i> and <i>geneIII</i> into the digested vector has been conducted. Thus a <i>mdnA-geneIII</i>-fusion part according to RFC25 was generated whereby <i>Age</i>I and <i>NgoM</i>IV overhangs are compatible and placed in frame with the protein sequence. The ligation of <i>Age</i>I and <i>NgoM</i>IV overhangs resulted in a scar coding for the threonine and glycine. Because the introduction of restriction sites before <i>mdnA</i> led to a great distance between ribosome binding site (RBS) and <i>mdnA</i>. A second RBS was inserted among <i>Sfo</i>I and <i>Xba</i>I recognition sites to ensure a sufficient expression rate of the <i>mdnA-geneIII</i>-fusion gene. The ''myc'' sequence located between <i>mdnA</i> and <i>gene III</i> allows the detection of the expression of the <i>mdnA-geneIII</i>-fusion protein on the surface of the phage using western blot or ELISA. In the last step the kanamycin resistance gene was disturbed because for phage display the selection of cells carriyng both helper phages and the phagemid is beneficial and the helper phages have a kanamycin resistance, too. Therefore a 300 bp fragment of the kanamycin resistance gene was deleted using the restriction enzyme <i>Nsi</i>I which had two recognition sites in the kanamycin gene. | ||
| Line 12: | Line 15: | ||
{| border="0" cellspacing="0" cellpadding="2" | {| border="0" cellspacing="0" cellpadding="2" | ||
| - | | [[File:UP cloning strategy.png|450px]] || [[File:UP pPDV089.png|350px]] | + | | [[File:UP cloning strategy.png|center|450px|thumb|'''Figure 1: Cloning strategy for creating a plasmid which can be used for phage display with <i>mdnA</i>. '''<i>MdnA</i> was cut out of the vector and a <i>mdnA-myc-geneIII</i> fusion gene was created and ligated with the vector pARW089 containing the <i>mdn</i>-cluster without <i>mdnA</i>.]] || [[File:UP pPDV089.png|center|350px|thumb|'''Figure 2: Designed vector pPDV089 carrying the <i>mdnA-myc-gene III</i> fusion gene.''' Therefore the <i>mdnA</i> sequence was cut out of the vector pARW089 and the ligated <i>mdnA-myc-geneIII</i> gene was inserted]] |
|- | |- | ||
| - | | align="center" | | + | | align="center" | |
| - | + | ||
|- | |- | ||
|} | |} | ||
| - | |||
| - | + | ===Control of expression of ''mdnA''-''myc''-''geneIII'' in ''E. coli''=== | |
| + | |||
| + | The expression of the ''mdnA''-''myc''-''geneIII'' fusion gene was analyzed by western blotting. ''E. coli'' cells transformed with the phagemid pPDV089 were harvested and lysated. The proteins were electrophoretically separated and transferred to a membrane. The ''mdnA''-''myc''-''geneIII''-fusion proteins were detected using specific anti-''myc''-antibodies and horseradish peroxidase (HRP)-linked antibodies as secondary antibodies. Enhanced chemiluminescence (ECL) was used to visualize the protein. ECL is based on the emission of light during the HRP-catalyzed oxidation of luminol, which was captured by a camera. The western blot analysis resulted in a band of a size just below the 30 kDa mark representing the ''mdnA''-''myc''-''geneIII''-fusion protein (24 kDa). | ||
| + | |||
| + | [[File:UP_coomassie+western blot.png|center|400px|thumb|'''Figure 3: Control of expression of ''mdnA''-''myc''-''geneIII'' in ''E. coli'' by western blotting.''' For detection anti-''myc''-antibodies and secondary HRP-linked antibodies were used. The resulting band represents the ''mdnA''-''myc''-''geneIII''-fusion protein.]] | ||
| + | <br> | ||
| + | <br> | ||
<br> | <br> | ||
| - | The next step was the detection of the expression of the ''mdnA-myc- | + | |
| - | For more precise results the absorption | + | ===Detection of phages carrying ''mdnA'' on their surface by ELISA=== |
| - | The graphic shows clearly the much higher absorption measured in wells, which were incubated with phage particles of interest produced in XL1-Blue cells. As has already pointed out this shows the succeeded expression of mdnA-c-myc- | + | |
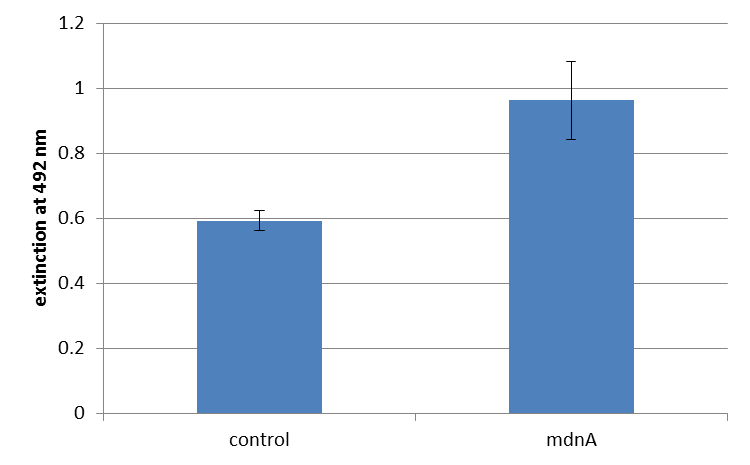
| + | The next step was the detection of the expression of the ''mdnA''-''myc''-''geneIII''-fusion gene on the surface of the phage. So ''E. coli'' cells strain XL1-Blue were first transformed with the phagemid pPDV089 before they were infected with helper phages. ''E. coli'' cells containing both plasmids were selected. An ELISA test was performed to determine whether these cells are able to produce phage particles carrying the MdnA peptide on their surface. To perform this test anti-''c-myc''-antibodies were immobilized on ELISA plates and incubated with purified phages. For detection a second antibody coupled with horse radish peroxidase (HRP) was used which binds the gene VIII protein of the phages. The HRP substrate o-phenyldiamine (OPD) was added and in case of binding a color reaction was expected. The color shift from achromatic to yellow in wells incubated with phages produced in XL1-Blue cells showed the successful expression of ''mdnA''-c-''myc''-''geneIII''-fusion protein on the phages.<br> | ||
| + | For more precise results the absorption at 492 nm was measured. The data were presented in a bar plot. As a negative control helper phages were added instead of produced phages. Furthermore two wells were prepared were the secondary antibody was not added. | ||
| + | The graphic shows clearly the much higher absorption measured in wells, which were incubated with phage particles of interest produced in XL1-Blue cells. As it has already been pointed out this shows the succeeded expression of ''mdnA''-c-''myc''-''geneIII''-fusion protein on the surface of the phages. | ||
<div align="center"> | <div align="center"> | ||
| - | [[File: | + | [[File:UP_ELISA3_mit_fehlerindikator.png|center|400px|thumb| '''Figure 4:''' Detection of phages carrying ''mdnA'' on their surface by ELISA. The bar plot shows the absorption at 492 nm. Anti-''myc''-antibodies were immobilized. For detection a second antibody coupled with horse radish peroxidase (HRP) was used which binds the gene VIII coat protein of the phages. The left bar shows the absorption of the wells containing helper phages (negative control), the right bar shows the absorption of wells containing ''mdnA'' carrying phages]]</div><br> |
| + | <br> | ||
| - | + | ===Testing phage display with unmodified ''mdnA'' to examine its suitability as screening method=== | |
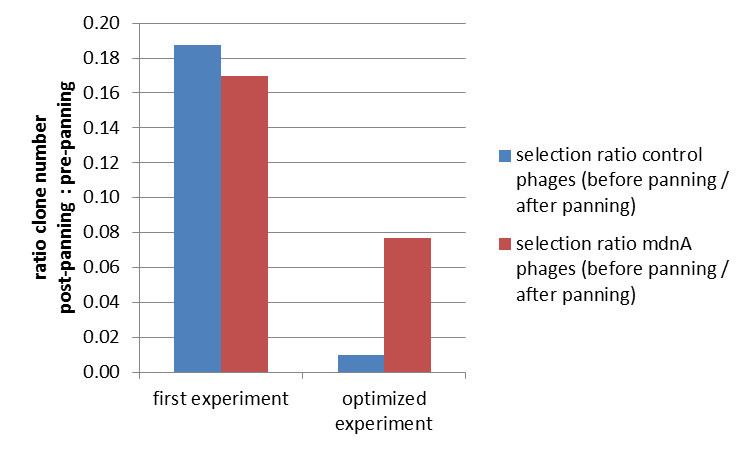
| + | To test the fundamental suitability of this screening method, phages representing unmodified ''mdnA'' on their surface and phages not representing ''mdnA'' (helper phages) in a ratio of one to one were incubated with immobilized trypsin which is known as a target of ''mdnA''. The display was conducted in ELISA plates. The bound phages were eluted using a buffer with low pH value and neutralized afterwards. To check how many phages interacted with trypsin, ''E. coli'' cells XL1-Blue were re-infected with eluted phages and plated on agar with different antibiotics. Cells infected with phages carrying ''mdnA'' are able to grow on agar with ampicillin whereas cells infected with helper phages are able to grow on agar with kanamycin. To control the success of the panning round additionally ''E. coli'' cells were infected with phage mix before panning and plated on agar. Subsequent the number of clones grew on ampicillin and kanamycin before and after panning was compared. During the running of this step it was noticed that much more cells were infected with helper phages than with phages carrying ''mdnA'' despite of the engaged 1:1 ratio. This was surprising and indicated that ''mdnA'' on the surface of the phages may inhibit their infectivity. After controlling the plates an infection ratio of phages carrying ''mdnA'' to helper phages of 1:400 was calculated. This fact should be analyzed in further experiments.<br> | ||
| + | The results of the first phage display are plotted in the figure below. After one panning round an enrichment of phages carrying ''mdnA'' was expected. This is attributable to the fact that phage particles carrying ''mdnA''-c-''myc'''-gene III-fusion protein on their surface are expected to bind specifically to the immobilized trypsin. Unfortunately this was not observed in this experiment. The ratio of cells growing on kanamycin agar before (4000) to cells growing on kanamycin agar (cells containing helper phages) after panning (750) was determined as 5:1. The ratio of cells growing on ampicillin agar before (12) to cells growing on ampicillin agar (cells containing ''mdnA'' carrying phages) after panning (2) was nearly equal. Thus no enrichment of ''mdnA'' carrying phages occurred in the first experiment. So it was decided to repeat this experiment under improved conditions. Therefor the number of washing steps during the described experimental procedure was increased. Here the ratio of cells growing on kanamycin agar before (3000) to cells growing on kanamycin agar (cells containing helper phages) after panning (29) was determined as 103:1. The ratio of cells growing on ampicillin agar before (26) to cells growing on ampicillin agar (cells containing ''mdnA'' carrying phages) after panning (2) was determined as 13:1. Thus an enrichment factor of eight was reached for the phages displaying ''mdnA'' on their surface. <br> | ||
| + | These results indicate that the unmodified ''mdnA'' expressed on the phages binds specifically to the immobilized trypsin. Therefore it can be deduced that ''mdnA'' is presented in a functional 3D structure. These findings suggest that phage display in general is an appropriate method for screening a recombinant ''mdnA'' library. Further experiments are required to optimize this system. | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | [[File:UP panning 3.png|center|400px|thumb| '''Figure 5: Optimization of phage display.''' After optimized conditions (right) a clear concentration of phages carrying ''mdnA'' after one panning round was noted. ''E. coli'' cells were infected with phage mix (helper phages and phages carrying ''mdnA'') before and after panning and plated on agar containing kanamycin or ampicillin. The ratio of cells growing on ampicillin or kanamycin agar before panning to cells growing on ampicillin or kanamycin agar after was calculated. Helper phages which acted as negative control have a kanamycine resistance whereby phages carrying ''mdnA'' have an ampicillin resistance.]] | ||
| + | |||
| + | ===Testing phage display with unmodified mdnA against further proteases=== | ||
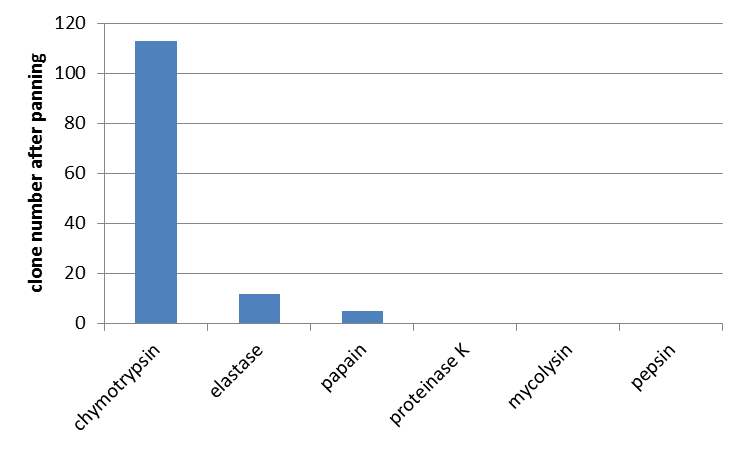
| + | |||
| + | Furthermore the interaction of unmodified mdnA with other proteases was determined. From the literature (Ziemert, 2010) the high inhibitory activity of microviridin L, besides trypsin, against chymotrypsin and elastase is also known. So a phage display with these enzymes was performed. Additionally papain, proteinase K, mycolysin and pepsin were tested for which no interaction was shown yet. For all enzymes an equal amount of ''E. coli'' cells and phages were used. In agreement with data from the literature interaction of microviridin with chymotrypsin and elastase was confirmed. Additionally an interaction with papain was noted. All other proteases were not bound by microviridin. | ||
| + | |||
| + | [[File:UP_test of different enzymes.png|center|400px|thumb|'''Figure 6: Phage display against different proteases.''' After panning the number of clones was counted. In agreement with data from the literature interaction of microviridin with chymotrypsin and elastase was confirmed. Additionally an interaction with papain was noted.]] | ||
| + | <br><br> | ||
| + | |||
| + | ===References=== | ||
| + | *Krebber, A., Bornhauser, S., Burmester, J., Honegger, A., Willuda, J., Bosshard, H. R., Plückthun, A. (1997) Reliable cloning of functional antibody variable domains from hybridomas and spleen cell repertoires employing a reengineered phage display system. J. Immunol. Meth. 201(1):35-55 | ||
| + | |||
| + | *Rakonjac J., Feng J., Model P. (1999). Filamentous phage are released from the bacterial membrane by a two-step mechanism involving a short C-terminal fragment of pIII. J Mol Biol. 289(5):1253-65 | ||
| + | *Smith, G.P. (1985). Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virus surface. Science 228: 1315-17 | ||
| + | |||
| + | *Ziemert, N., Ishida, K., Liaimer, A., Hertweck, C. & Dittmann, E. (2008). Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angewandte Chemie (International ed. in English) 47, 7756-9 | ||
| - | + | *Ziemert, N., Ishida, K., Weiz, A., Hertweck, C. & Dittmann, E. (2010). Exploiting the natural diversity of microviridin gene clusters for discovery of novel tricyclic depsipeptides. Applied and environmental microbiology 76, 3568-74 | |
| + | <br><br> | ||
Latest revision as of 03:33, 29 October 2011
Phage Display
Introduction
Phage Display is an efficient tool for selecting proteins or peptides with specific binding properties from a large recombinant library. In phage display proteins are presented on the surface of bacteriophages. This enables the coupling of phenotype and stable packaged genotype, because the proteins which form the phage including the proteins of interest are coded in its genome. To test the suitability of phage display system as an appropriate screening method for recombinant mdnA libraries a vector containing a mdnA-myc-geneIII-fusion gene was generated. This vector contains a plasmid origin of replication, so it can be amplified like plasmids. Additionally it contains a f1 ori which enables the packaging of single strand DNA into phages. The vector also contains the whole mdn-cluster which is needed to produce the MdnA peptide. Between mdnA and gene III is located a myc-tag, which is used for an easy detection. The successful expression of the MdnA-myc-geneIII-fusion protein on the surface of the phage was determined by ELISA using anti-myc antibody 9E10 after transforming E. coli cells and purifying the produced phages. The next step was to perform a phage display on a known target of the MdnA. To test the fundamental suitability of this screening method, phages presenting MdnA on their surface and phages not presenting MdnA in a ratio of one to one were incubated with immobilized trypsin which is known as a target of MdnA. After the first panning round a notable concentrating of phages carrying MdnA was recognized.
Cloning strategy
The phage display vector pPDV089 was derived from the plasmid pARW089 which carries the whole mdn-cluster. This plasmid contains a plasmid origin of replication and additionally a f1 ori which enables the packaging of single strand DNA into phages. For selective amplification ampcillin and kanamycin resistance genes are included. To create the phagemid pPDV089 standard cloning procedure were performed. First mdnA was excised using the restriction enzymes SfoI and AatII. The next step comprised the introduction of a mdnA-geneIII-fusion gene. Therefore gene III was amplified from pak100blaKDIR and mdnA from pARW089 by PCR. The primers were designed to enable the introduction of iGEM and other restriction sites required for further cloning steps. The purified PCR product geneIII was digested by NgoMIV and AatII whereas the PCR product mdnA was digested by SfoI and AgeI. Subsequent the three fragment ligation of mdnA and geneIII into the digested vector has been conducted. Thus a mdnA-geneIII-fusion part according to RFC25 was generated whereby AgeI and NgoMIV overhangs are compatible and placed in frame with the protein sequence. The ligation of AgeI and NgoMIV overhangs resulted in a scar coding for the threonine and glycine. Because the introduction of restriction sites before mdnA led to a great distance between ribosome binding site (RBS) and mdnA. A second RBS was inserted among SfoI and XbaI recognition sites to ensure a sufficient expression rate of the mdnA-geneIII-fusion gene. The myc sequence located between mdnA and gene III allows the detection of the expression of the mdnA-geneIII-fusion protein on the surface of the phage using western blot or ELISA. In the last step the kanamycin resistance gene was disturbed because for phage display the selection of cells carriyng both helper phages and the phagemid is beneficial and the helper phages have a kanamycin resistance, too. Therefore a 300 bp fragment of the kanamycin resistance gene was deleted using the restriction enzyme NsiI which had two recognition sites in the kanamycin gene.
Control of expression of mdnA-myc-geneIII in E. coli
The expression of the mdnA-myc-geneIII fusion gene was analyzed by western blotting. E. coli cells transformed with the phagemid pPDV089 were harvested and lysated. The proteins were electrophoretically separated and transferred to a membrane. The mdnA-myc-geneIII-fusion proteins were detected using specific anti-myc-antibodies and horseradish peroxidase (HRP)-linked antibodies as secondary antibodies. Enhanced chemiluminescence (ECL) was used to visualize the protein. ECL is based on the emission of light during the HRP-catalyzed oxidation of luminol, which was captured by a camera. The western blot analysis resulted in a band of a size just below the 30 kDa mark representing the mdnA-myc-geneIII-fusion protein (24 kDa).
Detection of phages carrying mdnA on their surface by ELISA
The next step was the detection of the expression of the mdnA-myc-geneIII-fusion gene on the surface of the phage. So E. coli cells strain XL1-Blue were first transformed with the phagemid pPDV089 before they were infected with helper phages. E. coli cells containing both plasmids were selected. An ELISA test was performed to determine whether these cells are able to produce phage particles carrying the MdnA peptide on their surface. To perform this test anti-c-myc-antibodies were immobilized on ELISA plates and incubated with purified phages. For detection a second antibody coupled with horse radish peroxidase (HRP) was used which binds the gene VIII protein of the phages. The HRP substrate o-phenyldiamine (OPD) was added and in case of binding a color reaction was expected. The color shift from achromatic to yellow in wells incubated with phages produced in XL1-Blue cells showed the successful expression of mdnA-c-myc-geneIII-fusion protein on the phages.
For more precise results the absorption at 492 nm was measured. The data were presented in a bar plot. As a negative control helper phages were added instead of produced phages. Furthermore two wells were prepared were the secondary antibody was not added.
The graphic shows clearly the much higher absorption measured in wells, which were incubated with phage particles of interest produced in XL1-Blue cells. As it has already been pointed out this shows the succeeded expression of mdnA-c-myc-geneIII-fusion protein on the surface of the phages.
Testing phage display with unmodified mdnA to examine its suitability as screening method
To test the fundamental suitability of this screening method, phages representing unmodified mdnA on their surface and phages not representing mdnA (helper phages) in a ratio of one to one were incubated with immobilized trypsin which is known as a target of mdnA. The display was conducted in ELISA plates. The bound phages were eluted using a buffer with low pH value and neutralized afterwards. To check how many phages interacted with trypsin, E. coli cells XL1-Blue were re-infected with eluted phages and plated on agar with different antibiotics. Cells infected with phages carrying mdnA are able to grow on agar with ampicillin whereas cells infected with helper phages are able to grow on agar with kanamycin. To control the success of the panning round additionally E. coli cells were infected with phage mix before panning and plated on agar. Subsequent the number of clones grew on ampicillin and kanamycin before and after panning was compared. During the running of this step it was noticed that much more cells were infected with helper phages than with phages carrying mdnA despite of the engaged 1:1 ratio. This was surprising and indicated that mdnA on the surface of the phages may inhibit their infectivity. After controlling the plates an infection ratio of phages carrying mdnA to helper phages of 1:400 was calculated. This fact should be analyzed in further experiments.
The results of the first phage display are plotted in the figure below. After one panning round an enrichment of phages carrying mdnA was expected. This is attributable to the fact that phage particles carrying mdnA-c-myc'-gene III-fusion protein on their surface are expected to bind specifically to the immobilized trypsin. Unfortunately this was not observed in this experiment. The ratio of cells growing on kanamycin agar before (4000) to cells growing on kanamycin agar (cells containing helper phages) after panning (750) was determined as 5:1. The ratio of cells growing on ampicillin agar before (12) to cells growing on ampicillin agar (cells containing mdnA carrying phages) after panning (2) was nearly equal. Thus no enrichment of mdnA carrying phages occurred in the first experiment. So it was decided to repeat this experiment under improved conditions. Therefor the number of washing steps during the described experimental procedure was increased. Here the ratio of cells growing on kanamycin agar before (3000) to cells growing on kanamycin agar (cells containing helper phages) after panning (29) was determined as 103:1. The ratio of cells growing on ampicillin agar before (26) to cells growing on ampicillin agar (cells containing mdnA carrying phages) after panning (2) was determined as 13:1. Thus an enrichment factor of eight was reached for the phages displaying mdnA on their surface.
These results indicate that the unmodified mdnA expressed on the phages binds specifically to the immobilized trypsin. Therefore it can be deduced that mdnA is presented in a functional 3D structure. These findings suggest that phage display in general is an appropriate method for screening a recombinant mdnA library. Further experiments are required to optimize this system.
Testing phage display with unmodified mdnA against further proteases
Furthermore the interaction of unmodified mdnA with other proteases was determined. From the literature (Ziemert, 2010) the high inhibitory activity of microviridin L, besides trypsin, against chymotrypsin and elastase is also known. So a phage display with these enzymes was performed. Additionally papain, proteinase K, mycolysin and pepsin were tested for which no interaction was shown yet. For all enzymes an equal amount of E. coli cells and phages were used. In agreement with data from the literature interaction of microviridin with chymotrypsin and elastase was confirmed. Additionally an interaction with papain was noted. All other proteases were not bound by microviridin.
References
- Krebber, A., Bornhauser, S., Burmester, J., Honegger, A., Willuda, J., Bosshard, H. R., Plückthun, A. (1997) Reliable cloning of functional antibody variable domains from hybridomas and spleen cell repertoires employing a reengineered phage display system. J. Immunol. Meth. 201(1):35-55
- Rakonjac J., Feng J., Model P. (1999). Filamentous phage are released from the bacterial membrane by a two-step mechanism involving a short C-terminal fragment of pIII. J Mol Biol. 289(5):1253-65
- Smith, G.P. (1985). Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virus surface. Science 228: 1315-17
- Ziemert, N., Ishida, K., Liaimer, A., Hertweck, C. & Dittmann, E. (2008). Ribosomal synthesis of tricyclic depsipeptides in bloom-forming cyanobacteria. Angewandte Chemie (International ed. in English) 47, 7756-9
- Ziemert, N., Ishida, K., Weiz, A., Hertweck, C. & Dittmann, E. (2010). Exploiting the natural diversity of microviridin gene clusters for discovery of novel tricyclic depsipeptides. Applied and environmental microbiology 76, 3568-74
 "
"