|
|
| (34 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| - | {{:Team:ETH Zurich/Templates/Header/Overview|currPage=CircuitDesign}} | + | {{:Team:ETH Zurich/Templates/HeaderNew}} |
| - | {|class="roundContainer"
| + | |
| - | |style="font-size:2em; height: 30px" class="overview"|Circuit Design
| + | |
| - | |-
| + | |
| - | |'''To realize the smoke sensor (see [[Team:ETH_Zurich/Overview/Project|Project Idea]]), we implemented two distinct network designs: one SmoColi sensing acethaldehyde (see [[#Signal Detection|Figure 1]]) and one sensing xylene (see [[#Signal Detection|Figure 2]]). Both designs share the same signal pre-processing, transduction and processing units, and a similar design in the signal detection unit. '''
| + | |
| - | |}
| + | |
| | | | |
| - | {|class="roundContainer" | + | {{:Team:ETH Zurich/Templates/SectionStart}} |
| - | |
| + | = SmoColi - Circuit Design = |
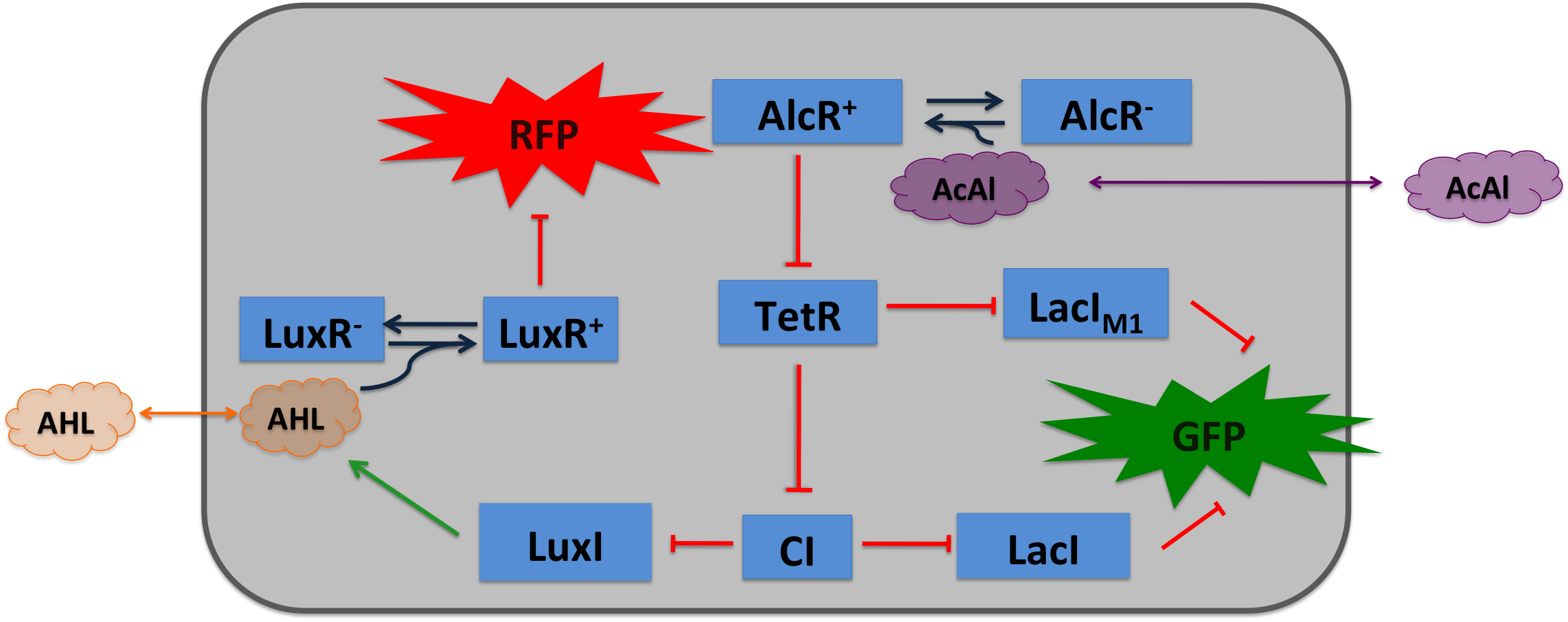
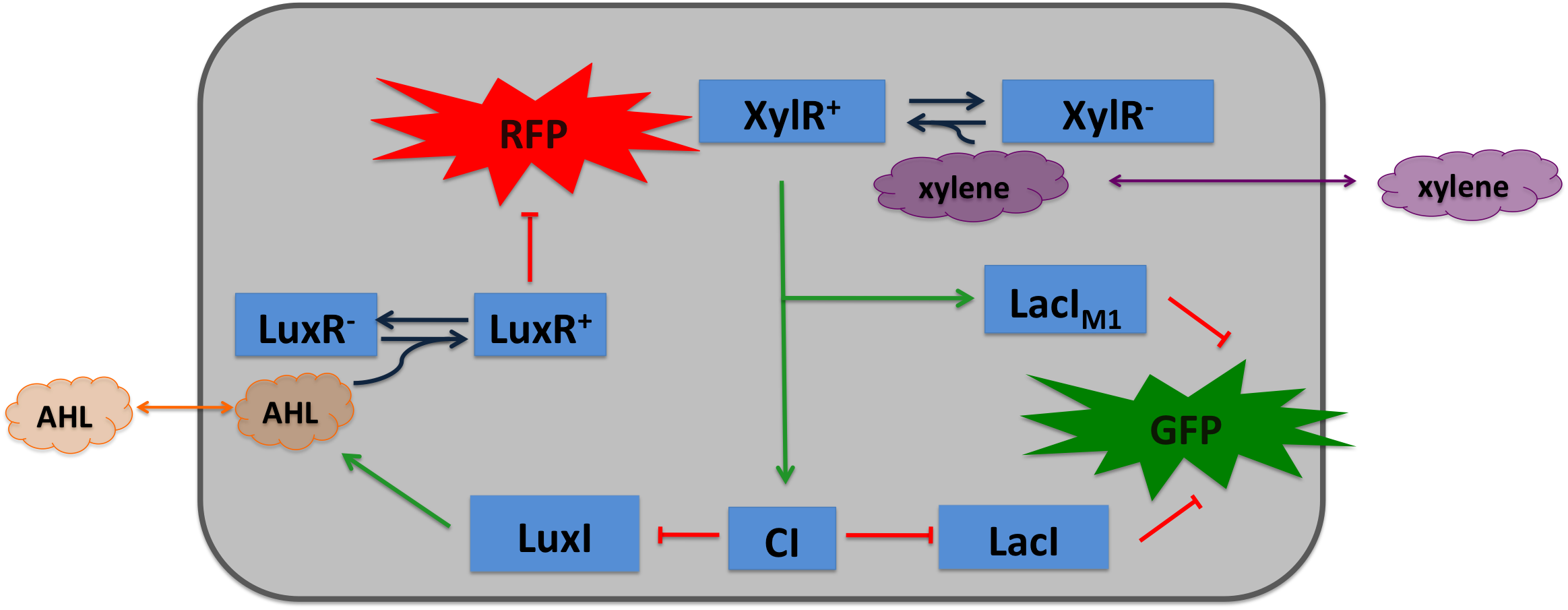
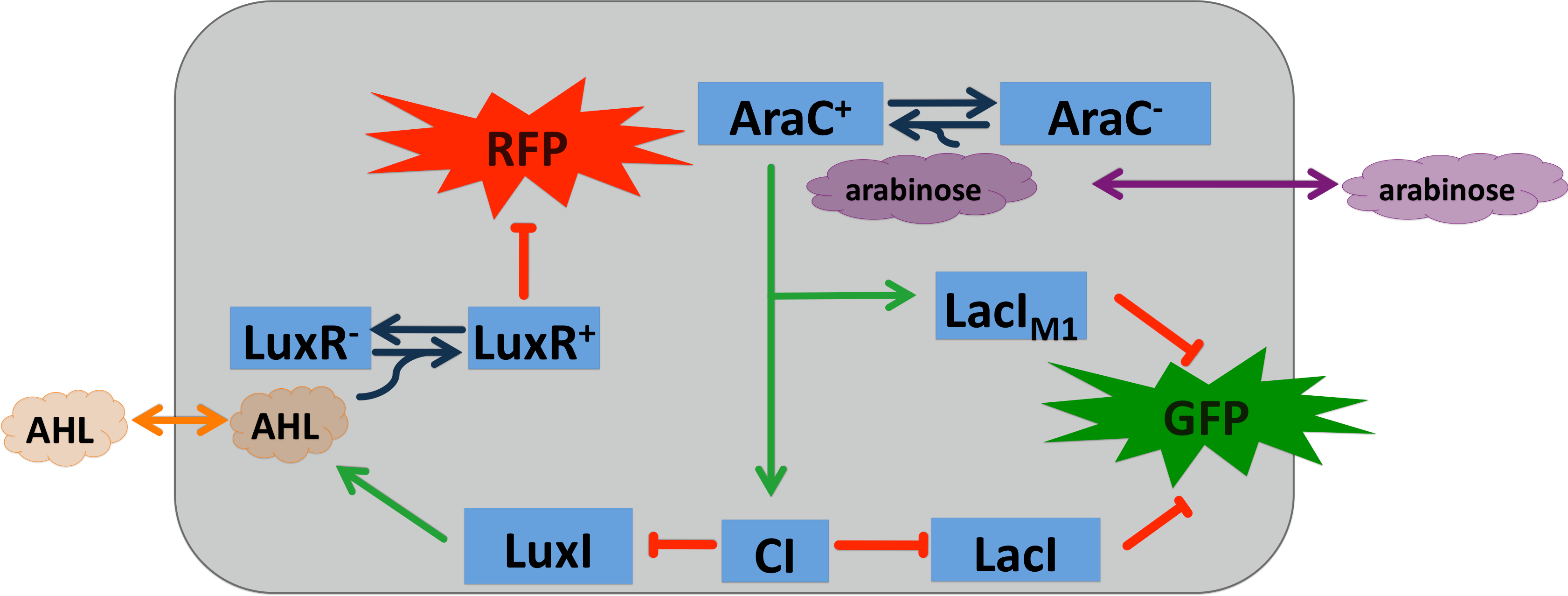
| - | === Signal Pre-Processing === | + | To realize the smoke sensor (see [[Team:ETH_Zurich/Overview/Project|Project Idea]]), we implemented three distinct network designs: one SmoColi sensing acethaldehyde (see Figure 1), one sensing xylene (see Figure 2) and one sensing arabinose (see Figure 3). All designs share the same '''signal pre-processing''', '''processing units''', and a similar design in the '''signal detection unit'''. |
| - | The first step for both designs is to map an absolute acetaldehyde (xylene) concentration into a spatial signal for further analysis. For this pre-processing step we constructed a flow channel and placed the cells therein. Initially the flow-channel is filled with a medium containing no acetaldehyde (xylene). The channel is then connected at one side to the medium for which the acetaldehyde (xylene) concentration should be measured. Acetaldehyde (xylene) diffuses from this medium into the channel.
| + | |
| | | | |
| - | Acetaldehyde is naturally degraded in E. coli by the acetaldehyde dehydrogenase. For the second design we engineered SmoColi to also degrade xylene by integrating the upper TOL pathway into the cells. The degradation of acetaldehyde (xylene) counteracts its diffusion into the channel, leading to the establishment of a spatial steady state acetaldehyde (xylene) concentration gradient. The shape and hight of the gradient only depends on the cell density, the degradation rate per cell, the diffusion constant, and the acetaldehyde (xylene) concentration in the medium to measure. Thus, by increasing or decreasing the acetaldehyde (xylene) concentration in the medium to measure and keeping the other parameters unchanged, the gradient is shifted to the right or left. In the following we describe how we use this spatial shift of the gradient to detect and quantify the initial concentration of acetaldehyde (xylene) in the medium.
| + | {|style="border: none;" align="center" |
| - | | + | |valign="top"|[[File:Design2.png|450px|center|thumb|'''Figure 1: Molecular mechanism of SmoColi with acetaldehyde sensor.''' ]] |
| - | === Signal Detection === | + | |valign="top"|[[File:Alternative_Xylene_system.png|450px|center|thumb|'''Figure 2: Molecular mechanism of SmoColi with xylene sensor.''']] |
| - | [[File:Design2.png|400px|left|thumb|'''Figure 1: Molecular mechanism of SmoColi with acetaldehyde sensor ''' ]] | + | |
| - | [[File:Alternative_Xylene_system.png|400px|right|thumb|'''Figure 2: Molecular mechanism of SmoColi with xylene sensor.''' ]] | + | |
| - | <br clear="all" />
| + | |
| - | For the first design (second design), the small signaling molecule acetaldehyde (xylene) is supplied in the extra-cellular medium. Acetaldehyde (xylene) can diffuse through cellular membranes and associates with its constitutively expressed receptor AlcR (XylR). Upon binding, the AlcR-acetaldehyde (XylR-xylene) complex can bind to its cognate operator, and inhibits (activates) the transcription of the genes under the control of the P<sub>AlcR</sub> (P<sub>U</sub>) promoter.
| + | |
| - |
| + | |
| - | Since for the further signal processing it was necessary to have a POPS signal proportional to the signaling molecule concentration, and since the acetaldehyde sensor inhibits transcription, we decided to include an additional unit to invert the transduced signal downstream of acetaldehyde (this element is not necessary for the xylene sensor): we put TetR under the transcriptional control of AlcR, and since TetR acts as a transcriptional inhibitor for downstream proteins, the transduced acetaldehyde signal has a similar qualitative input/output relation as the transduced xylene sensor without inverter.
| + | |
| - | | + | |
| - | === Signal Processing I: the Band-Pass Filter ===
| + | |
| - | [[File:ETH_states_of_bandpass.png|300px|right|thumb|'''Figure 3: Circuit operations for SmoColi''' exposed to medium, low and zero xylene concentration resulting in a GFP band. Triangles indicating the gradient of AHL (blue) and sensor molecule (violett), receptively. Non active interactions and non expressed Proteins are indicated light colors.]]
| + | |
| - | | + | |
| - | To realize the spatial green fluorescence band indicating the acetaldehyde (xylene) concentration, we implemented a signal processing unit, which transduces the incoming signal if and only if the signal is neither too strong nor too weak (band-pass filter [[#Ref1|[1]]]):
| + | |
| - | | + | |
| - | TetR represses (XylR enhances) the expression of the LacI<sub>M1</sub> repressor (codon-modified LacI) and the lambda repressor CI. Thus, high acetaldehyde (xylene) concentrations result in high cytoplasmic levels of CI and of LacI<sub>M1</sub>. Since LacI<sub>M1</sub> represses the expression of the green fluorescent protein (GFP), no green fluorescence signal can be detected for high concentrations.
| + | |
| - | | + | |
| - | Low acetaldehyde (xylene) concentrations result in low LacI<sub>M1</sub> and CI concentrations. Although in this case, LacI<sub>M1</sub> does not repress GFP production, GFP is transcriptional repressed by the non-codon modified LacI, which is expressed at low CI concentrations. Only intermediate signals results in a moderate level of CI and LacI<sub>M1</sub>. The repression of CI is significantly higher than the one of LacI<sub>M1</sub>. While moderate levels of CI effectively shuts off LacI expression, the LacI<sub>M1</sub> concentration is to low to repress GFP
| + | |
| - | <br clear ="all" />
| + | |
| - | === Signal Processing II: the Alarm Signal ===
| + | |
| - | [[File:ETH_Setup.png|300px|right|thumb|'''Figure 4: Circuit operations for SmoColi''' exposed to medium, low and zero xylene concentration resulting in a GFP band. Triangles indicating the gradient of AHL (blue) and sensor molecule (violett), receptively. Non active interactions and non expressed Proteins are indicated light colors.]]
| + | |
| - | | + | |
| - | To realize the alarm signal for toxic acetaldehyde (xylene) concentrations, we put the N-Acyl homoserine lactone (AHL) Synthase LuxI under the transcriptional control of CI, which is thus expressed only at low acetaldehyde (xylene) concentrations. LuxI syntesizes the small signaling molecule AHL which can quickly diffuse through the cell membrane in the extracellular space. Due to its relatively high diffusion constant, AHL diffuses through the whole tube and, thus, is present even in cells where no AHL is produced. Inside a cell, AHL can bind to its constitutively expressed receptor protein LuxR, and the thus activated LuxR represses the transcription of a red fluorescent protein (RFP). Thus, if the acetaldehyde concentration is below a threshold at least for a large enough minority of the cells in the channel, RFP is repressed in all cells. Only if the acetaldehyde concentration is above the threshold for all cells, no AHL is present in the channel and RFP is produced. Thus, the alarm signal RFP can be used to signal critical and potentially dangerous acetaldehyde (xylene) concentrations.
| + | |
| | |} | | |} |
| | + | [[File:Arabinose system.png|450px|center|thumb|'''Figure 3: Circuit operations of SmoColi with arabinose sensor''' ]] |
| | | | |
| - | {|class="roundContainer"
| + | For a detailed '''explanation''' about how the circuit works [[Team:ETH_Zurich/Overview/Informationprocessing#Signal_Processing |click here]]. |
| - | | | + | |
| | | | |
| - | == References ==
| + | {{:Team:ETH Zurich/Templates/SectionEnd}} |
| - | <span id="Ref1">[1] [http://www.nature.com/nature/journal/v434/n7037/full/nature03461.html Subhayu Basu, Yoram Gerchman1, Cynthia H. Collins, Frances H. Arnold & Ron Weiss: '''A synthetic multicellular system for programmed pattern formation''', Nature 2005, 434: 1130-11342]</span>
| + | |
| | | | |
| - | |}
| + | {{:Team:ETH Zurich/Templates/HeaderNewEnd}} |
 "
"