Team:Potsdam Bioware/Methods
From 2011.igem.org
KatharinaB (Talk | contribs) (→Methods) |
(→Methods) |
||
| (37 intermediate revisions not shown) | |||
| Line 3: | Line 3: | ||
== Methods== | == Methods== | ||
| - | ''' | + | === ''Established Methods'' === |
| - | + | ||
| - | + | ||
| + | ===Cloning=== | ||
| + | ====Polymerase Chain Reaction==== | ||
| - | + | The purpose of PCR is to amplify a gene to get a huge number of copies of this. The sample is denaturated at 94°C. The resulting single strands give the possibility of primer binding. The primers bind to their complementary strand at a specific annealing temperature. Due to that, the polymerase can start the elongating of the primer and amplifying the template DNA. | |
| - | + | ||
| - | + | ||
| + | ====PCR purification==== | ||
| - | + | If the PCR conditions are well optimized, most of the added primers and dNTPs will be used up during the PCR amplification. For further cloning steps you need to have only one major PCR band visible on your agarose gel. To get rid of interfering factors you need to purify the PCR product. For purification we used the NucleoSpin Extract II Kit from Macherey-Nagel (Düren) as well as the Wizard<sup>®</sup> SV Gel and PCR Clean-Up System from Promega (Darmstadt). | |
| - | < | + | |
| - | Phage display is a powerful tool for | + | |
| - | In M13 phage display genes from a DNA-library are usually fused to the gene III coat protein which occurs in five. The gene III protein is responsible for phage infection and releasing phage particles after production (Holliger and Riechmann, 1997; Stengele et al., 1990). Along with insertion of the gene of interest into the phage genome (McCafferty, 1990) phagemid vectors are established (Mead, 1988). Phagemids contain phage and bacterial origins of replication, the gene encoding the coat protein for fusion and a marker gene. To produce functional phages, co-infection with helper phages is required. These plasmids contain all genes used for producing phage particles. Its genome carries a mutation which leads to more efficient packaging of the phage genome of interest in relation to the helper plasmid genome.<br> | + | ====Vector dephosphorylation==== |
| + | |||
| + | Before ligation we dephosphorylated our vectors to diminish the rate of religation. Therefore we used the TSAP Thermosensitive Alkaline Phosphatase from Promega (Darmstadt) according to manufacturers protocol. | ||
| + | |||
| + | |||
| + | ====Ligation==== | ||
| + | |||
| + | DNA ligation is a process of linking complementary overhangs of DNA molecules, which were created by restriction enzyme digestion. The ligase is a enzyme which connects the ends via covalent phosphodiester bonds between 3´hydroxyl ends of one nucleotide and 5´phosphate ends of the other fragment. For our experiments we used the T4-DNA-Ligase. | ||
| + | |||
| + | |||
| + | ====Competent cells==== | ||
| + | |||
| + | Competent cells are one of the most important tools in molecular biology. Compared to normal cells, the competent cells got a higher chance to include new generated vectors. After transformation of these new organisms with the generated plasmid, they got new abilities owing to the used plasmid. | ||
| + | In this lab we used competent ''E.coli'' of the strains XL1-Blue, ER2738 and RV308. The competent cells itself were produced by using rubidium chloride. The normal cells resistance was tested by using LB plates with several antibiotics. Additional the ability of transformation was tested by using the pUK–vector. | ||
| + | |||
| + | |||
| + | ====Transformation==== | ||
| + | |||
| + | The aim of transformations is to introduce a foreign plasmid into competent bacteria cells to amplify the plasmid. We used a protocol for heat shock transformation. The DNA is forced into the cells by incubating cells and DNA on ice, putting them on 42°C and putting them back on ice. The bacteria is caused to take in the DNA. Afterwards the cells are plated on LB media, which contain appropriate antibiotica. | ||
| + | |||
| + | ====Fluorescence spectroscopy==== | ||
| + | |||
| + | We checked the functionality and performance of our promoters by using the spectrofluorometer Jasco FP-6500. In this process we excitated the sample for checking of YFP by 500 nm and measured the resultant emission between 510 and 580 nm. By checking the emission of CFP we excitated the sample by 433 nm and measured the resultant emission between 445 and 520 nm. | ||
| + | |||
| + | |||
| + | ====Fillin-reactions==== | ||
| + | |||
| + | A fill-in reaction is a common method for producing blunt-ended DNA fragments by filling 5´overhangs. The enzyme used for this reaction is the large subunit of the ''E. coli'' DNA polymerase (Klenow fragment). | ||
| + | |||
| + | |||
| + | ====DNA gel electrophoresis==== | ||
| + | |||
| + | Before , e.g. a PCR product is used in further applications, it has to be checked if there is a product. Agarose gel electrophoresis is the easiest way of separating and analyzing DNA. The DNA is visualized in the gel by GelRed. This binds strongly to the DNA by intercalating between the bases. Using UV light it starts to fluorescence. The DNA is mixed with loading dye, which makes it easy to load the sample. For comparing fragments with known size with fragments of unknown size a ladder is used. For separating our samples we used 0.8%-1.5% agarose gels. | ||
| + | |||
| + | |||
| + | ====Agarose gel extraction==== | ||
| + | |||
| + | This method requires to cut out the band which matches the expected size from an agarose gel. The gel-slice needs to be dissolved. The solution is applied to a spin-column, which is washed afterwards. The DNA can then be eluted in a small volume of buffer or water. We used the NucleoSpin Extract II Kit from Macherey-Nagel (Düren). The purified samples can then be used for further cloning applications. | ||
| + | |||
| + | |||
| + | ====Random mutagenesis==== | ||
| + | |||
| + | Random mutagenesis is an important application in protein engineering, e.g. in the design of proteins with novel features. We used error-prone-PCR, where you try to hinder the polymerase with additionally high amounts of Mg<sup>2+</sup> and Mn<sup>2+</sup> . | ||
| + | |||
| + | |||
| + | ====Miniprep==== | ||
| + | |||
| + | Plasmid DNA extraction involves the separation of the plasmid from the genomic DNA. The cell wall needs to be lysed and the plasmid and genomic DNA to be denaturated, which is done with a buffer containing SDS and sodium hydroxide. In the next step the plasmid has to be renatured using a neutralizing buffer. For plasmid DNA minipreparation we used the NucleoSpin Plasmid Kit from Macherey-Nagel (Düren) according to the manufacturers protocol. | ||
| + | |||
| + | |||
| + | ====Midiprep==== | ||
| + | Plasmid DNA midiprep is done for large scale plasmid DNA isolation. | ||
| + | |||
| + | ====ELISA==== | ||
| + | Enzyme-linked immunosorbent assay (ELISA) is a widely spread method to detect protein-protein interactions through an antigene- antibody reaction. In our project it was used to detect the expressed mdnA-myc-gene III on the surface of phages. | ||
| + | |||
| + | ====Maldi-TOF==== | ||
| + | |||
| + | Mass spectrometry is an excellent technique which allows the measurement of mass to charge ratios of biomolecules. In case of MALDI-TOF (matrix associated laser desorption/ionization-time of flight) the sample is deposited on a matrix and then irradiated with a strong UV-laser beam. This leads to evaporation and ionization of matrix and sample molecules. The vaporized and ionized sample molecules are then accelerated in an electric field. The speed of the flying particles is proportional to m^-0.5 (m=mass of particle) thus heavy particles reach the detector later than lighter particles. In the reflector mode the flying particles reach the end of the evacuated channel, where their flying direction is reversed. This leads to an improved resolution of yielded particles. | ||
| + | |||
| + | |||
| + | |||
| + | ====Cloning of BioBricks==== | ||
| + | |||
| + | All the genes for BioBrick production were amplified by PCR. Due to the incorporation of the sequences of the iGEM restriction enzymes into the primer sequence our BioBricks comply with the BioBrick standards. We used the <i>E. coli</i> strain XL1-Blue as host for cloning of all parts. The sequence of the final constructed BioBrick was verified by sequencing. | ||
| + | |||
| + | |||
| + | ====HPLC analysis==== | ||
| + | |||
| + | HPLC is used in analysis for identifying and purifying individual compounds in a mixture. We wanted to purify the microviridin for further analysis. After incubation of an <i>E. coli</i> culture for reaching an OD600 between 0.5 and 0.6 the cells were centrifuged and the pellet was washed with Tris-HCl. The pellet was resuspended in H<sub>2</sub>O. The cells were then lysed via sonication and centrifuged. The supernatant was transferred to Sep-Pak Plus C18 Cartridges. After washing the samples were eluted from the cartridge in methanol and put in a speedvac. Afterwards the remaining pellet was resuspended in methanol, filtered and put on a HPLC vial. HPLC analyses were carried out on a Shimadzu HPLC. Fractions were sampled by hand. | ||
| + | |||
| + | ====Principle of phage display==== | ||
| + | <br> | ||
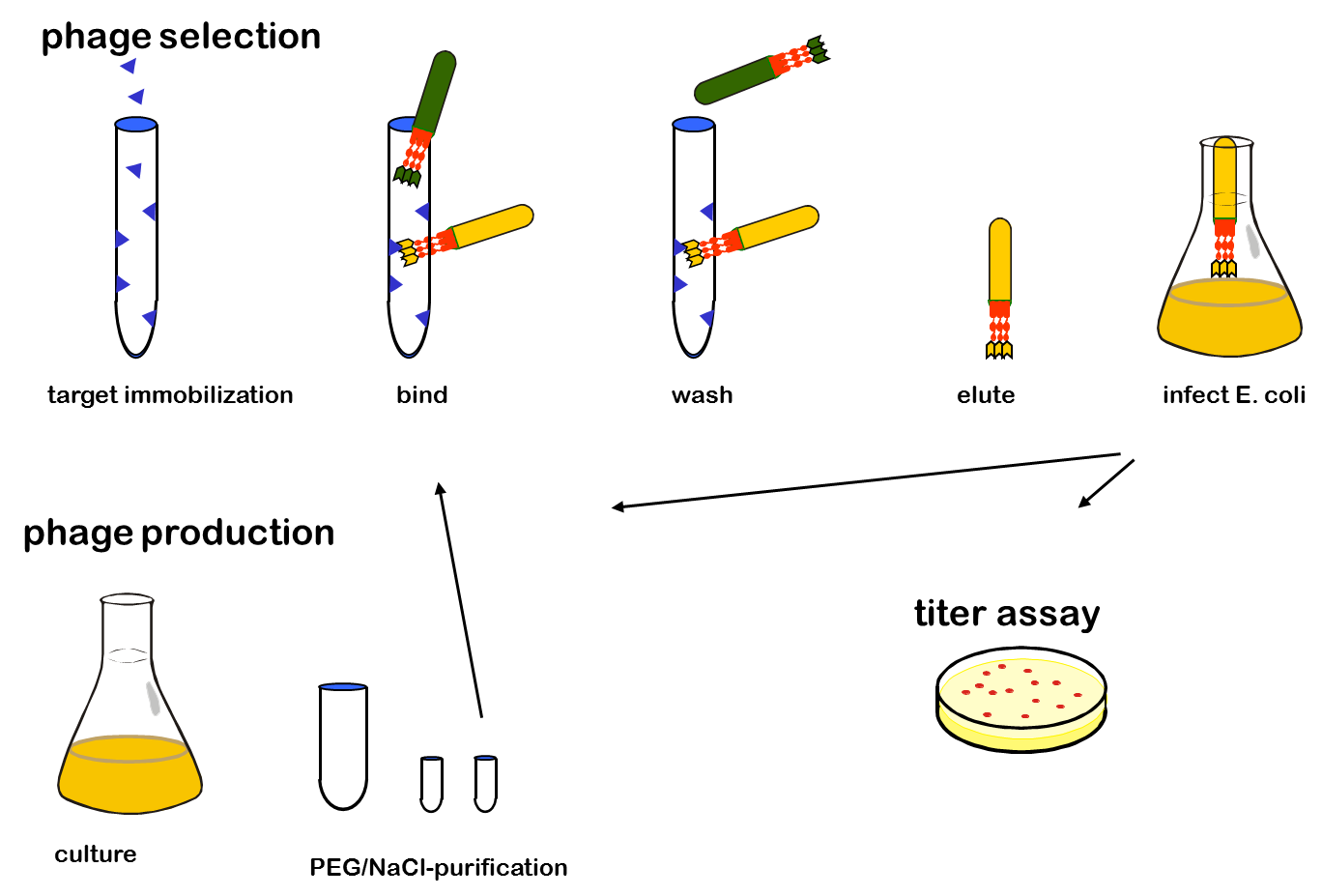
| + | Phage display is a powerful tool for selecting peptides or proteins that bind and regulate the function of target proteins. It is defined as a system in which the protein and its encoding gene are physically linked. The coupling of peptides to phages and the subsequent selection of binders was first described by George P. Smith in 1985. The bacterial viruses principally used are members of the single-stranded filamentous phage family M13. After attachment to the bacterial F-pilus and entering the cells this phages stripe of their protein coat and convert their DNA in the double stranded form. Than replication, expression and assembly of phage particles occurs. About 200 phages can be produced per cell and generation. The M13 phage is a non-lytic phage, that means that they can be released from bacterial cells without causing cell death.<br> | ||
| + | In M13 phage display genes from a DNA-library are usually fused to the gene III coat protein which occurs in five. The gene III protein is responsible for phage infection and releasing phage particles after production (Holliger and Riechmann, 1997; Stengele et al., 1990). Along with insertion of the gene of interest into the phage genome (McCafferty et al., 1990) phagemid vectors are established (Mead, 1988). Phagemids contain phage and bacterial origins of replication, the gene encoding the coat protein for fusion and a marker gene. To produce functional phages, co-infection with helper phages is required. These plasmids contain all genes used for producing phage particles. Its genome carries a mutation which leads to more efficient packaging of the phage genome of interest in relation to the helper plasmid genome.<br> | ||
One advantage of phage display is the rapid identification and concentration of target binding proteins through a procedure called biopanning. Bound phages are enriched by consecutive cycles of incubation, washing, amplification and selection. Therefor the target molecules are immobilized on ELISA plates or immune tubes. After an appropriate number of panning rounds, the inserts from the enriched phage can be sequenced to identify the interacting proteins.<br> | One advantage of phage display is the rapid identification and concentration of target binding proteins through a procedure called biopanning. Bound phages are enriched by consecutive cycles of incubation, washing, amplification and selection. Therefor the target molecules are immobilized on ELISA plates or immune tubes. After an appropriate number of panning rounds, the inserts from the enriched phage can be sequenced to identify the interacting proteins.<br> | ||
<br> | <br> | ||
| - | [[File:UP Filamentous phage.png|center|500px|thumb|'''Figure: '''Filamentous phage displaying a protein of interest ]]<br> | + | [[File:UP Filamentous phage.png|center|500px|thumb|'''Figure 1: '''Filamentous phage displaying a protein of interest ]]<br> |
| + | |||
| + | |||
| + | [[File:UP Procedere.png|center|400px|thumb|'''Figure 2: '''Phage display procedure ]]<br> | ||
| + | <br> | ||
| + | References:<br> | ||
| + | <br> | ||
| + | Holliger P., Riechmann L. (1997) A conserved infection pathway for filamentous bacteriophages is suggested by the structure of the membrane penetration domain of the minor coat protein g3p from phage fd. Structure 5(2):265-75. | ||
| + | |||
| + | McCafferty J., Griffiths A.D., Winter G., Chiswell D.J. (1990) Phage antibodies: Filamentous phage displaying antibody variable domains. Nature 348: 552-554 | ||
| + | |||
| + | Mead DA, Kemper B (1988) Chimeric single stranded DNA phage-plasmid cloning vectors. Biotchnology 10: 85-102 | ||
| + | Smith G.P. (1985) Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virus surface. Science 228: 1315-1317 | ||
| - | + | Stengele I., Bross P., Garcés X., Giray J., Rasched I. (1990) Dissection of functional domains in phage fd adsorption protein. Discrimination between attachment and penetration sites. J Mol Biol. Mar 5;212(1):143-9. | |
Latest revision as of 03:43, 22 September 2011
Methods
Established Methods
Cloning
Polymerase Chain Reaction
The purpose of PCR is to amplify a gene to get a huge number of copies of this. The sample is denaturated at 94°C. The resulting single strands give the possibility of primer binding. The primers bind to their complementary strand at a specific annealing temperature. Due to that, the polymerase can start the elongating of the primer and amplifying the template DNA.
PCR purification
If the PCR conditions are well optimized, most of the added primers and dNTPs will be used up during the PCR amplification. For further cloning steps you need to have only one major PCR band visible on your agarose gel. To get rid of interfering factors you need to purify the PCR product. For purification we used the NucleoSpin Extract II Kit from Macherey-Nagel (Düren) as well as the Wizard® SV Gel and PCR Clean-Up System from Promega (Darmstadt).
Vector dephosphorylation
Before ligation we dephosphorylated our vectors to diminish the rate of religation. Therefore we used the TSAP Thermosensitive Alkaline Phosphatase from Promega (Darmstadt) according to manufacturers protocol.
Ligation
DNA ligation is a process of linking complementary overhangs of DNA molecules, which were created by restriction enzyme digestion. The ligase is a enzyme which connects the ends via covalent phosphodiester bonds between 3´hydroxyl ends of one nucleotide and 5´phosphate ends of the other fragment. For our experiments we used the T4-DNA-Ligase.
Competent cells
Competent cells are one of the most important tools in molecular biology. Compared to normal cells, the competent cells got a higher chance to include new generated vectors. After transformation of these new organisms with the generated plasmid, they got new abilities owing to the used plasmid. In this lab we used competent E.coli of the strains XL1-Blue, ER2738 and RV308. The competent cells itself were produced by using rubidium chloride. The normal cells resistance was tested by using LB plates with several antibiotics. Additional the ability of transformation was tested by using the pUK–vector.
Transformation
The aim of transformations is to introduce a foreign plasmid into competent bacteria cells to amplify the plasmid. We used a protocol for heat shock transformation. The DNA is forced into the cells by incubating cells and DNA on ice, putting them on 42°C and putting them back on ice. The bacteria is caused to take in the DNA. Afterwards the cells are plated on LB media, which contain appropriate antibiotica.
Fluorescence spectroscopy
We checked the functionality and performance of our promoters by using the spectrofluorometer Jasco FP-6500. In this process we excitated the sample for checking of YFP by 500 nm and measured the resultant emission between 510 and 580 nm. By checking the emission of CFP we excitated the sample by 433 nm and measured the resultant emission between 445 and 520 nm.
Fillin-reactions
A fill-in reaction is a common method for producing blunt-ended DNA fragments by filling 5´overhangs. The enzyme used for this reaction is the large subunit of the E. coli DNA polymerase (Klenow fragment).
DNA gel electrophoresis
Before , e.g. a PCR product is used in further applications, it has to be checked if there is a product. Agarose gel electrophoresis is the easiest way of separating and analyzing DNA. The DNA is visualized in the gel by GelRed. This binds strongly to the DNA by intercalating between the bases. Using UV light it starts to fluorescence. The DNA is mixed with loading dye, which makes it easy to load the sample. For comparing fragments with known size with fragments of unknown size a ladder is used. For separating our samples we used 0.8%-1.5% agarose gels.
Agarose gel extraction
This method requires to cut out the band which matches the expected size from an agarose gel. The gel-slice needs to be dissolved. The solution is applied to a spin-column, which is washed afterwards. The DNA can then be eluted in a small volume of buffer or water. We used the NucleoSpin Extract II Kit from Macherey-Nagel (Düren). The purified samples can then be used for further cloning applications.
Random mutagenesis
Random mutagenesis is an important application in protein engineering, e.g. in the design of proteins with novel features. We used error-prone-PCR, where you try to hinder the polymerase with additionally high amounts of Mg2+ and Mn2+ .
Miniprep
Plasmid DNA extraction involves the separation of the plasmid from the genomic DNA. The cell wall needs to be lysed and the plasmid and genomic DNA to be denaturated, which is done with a buffer containing SDS and sodium hydroxide. In the next step the plasmid has to be renatured using a neutralizing buffer. For plasmid DNA minipreparation we used the NucleoSpin Plasmid Kit from Macherey-Nagel (Düren) according to the manufacturers protocol.
Midiprep
Plasmid DNA midiprep is done for large scale plasmid DNA isolation.
ELISA
Enzyme-linked immunosorbent assay (ELISA) is a widely spread method to detect protein-protein interactions through an antigene- antibody reaction. In our project it was used to detect the expressed mdnA-myc-gene III on the surface of phages.
Maldi-TOF
Mass spectrometry is an excellent technique which allows the measurement of mass to charge ratios of biomolecules. In case of MALDI-TOF (matrix associated laser desorption/ionization-time of flight) the sample is deposited on a matrix and then irradiated with a strong UV-laser beam. This leads to evaporation and ionization of matrix and sample molecules. The vaporized and ionized sample molecules are then accelerated in an electric field. The speed of the flying particles is proportional to m^-0.5 (m=mass of particle) thus heavy particles reach the detector later than lighter particles. In the reflector mode the flying particles reach the end of the evacuated channel, where their flying direction is reversed. This leads to an improved resolution of yielded particles.
Cloning of BioBricks
All the genes for BioBrick production were amplified by PCR. Due to the incorporation of the sequences of the iGEM restriction enzymes into the primer sequence our BioBricks comply with the BioBrick standards. We used the E. coli strain XL1-Blue as host for cloning of all parts. The sequence of the final constructed BioBrick was verified by sequencing.
HPLC analysis
HPLC is used in analysis for identifying and purifying individual compounds in a mixture. We wanted to purify the microviridin for further analysis. After incubation of an E. coli culture for reaching an OD600 between 0.5 and 0.6 the cells were centrifuged and the pellet was washed with Tris-HCl. The pellet was resuspended in H2O. The cells were then lysed via sonication and centrifuged. The supernatant was transferred to Sep-Pak Plus C18 Cartridges. After washing the samples were eluted from the cartridge in methanol and put in a speedvac. Afterwards the remaining pellet was resuspended in methanol, filtered and put on a HPLC vial. HPLC analyses were carried out on a Shimadzu HPLC. Fractions were sampled by hand.
Principle of phage display
Phage display is a powerful tool for selecting peptides or proteins that bind and regulate the function of target proteins. It is defined as a system in which the protein and its encoding gene are physically linked. The coupling of peptides to phages and the subsequent selection of binders was first described by George P. Smith in 1985. The bacterial viruses principally used are members of the single-stranded filamentous phage family M13. After attachment to the bacterial F-pilus and entering the cells this phages stripe of their protein coat and convert their DNA in the double stranded form. Than replication, expression and assembly of phage particles occurs. About 200 phages can be produced per cell and generation. The M13 phage is a non-lytic phage, that means that they can be released from bacterial cells without causing cell death.
In M13 phage display genes from a DNA-library are usually fused to the gene III coat protein which occurs in five. The gene III protein is responsible for phage infection and releasing phage particles after production (Holliger and Riechmann, 1997; Stengele et al., 1990). Along with insertion of the gene of interest into the phage genome (McCafferty et al., 1990) phagemid vectors are established (Mead, 1988). Phagemids contain phage and bacterial origins of replication, the gene encoding the coat protein for fusion and a marker gene. To produce functional phages, co-infection with helper phages is required. These plasmids contain all genes used for producing phage particles. Its genome carries a mutation which leads to more efficient packaging of the phage genome of interest in relation to the helper plasmid genome.
One advantage of phage display is the rapid identification and concentration of target binding proteins through a procedure called biopanning. Bound phages are enriched by consecutive cycles of incubation, washing, amplification and selection. Therefor the target molecules are immobilized on ELISA plates or immune tubes. After an appropriate number of panning rounds, the inserts from the enriched phage can be sequenced to identify the interacting proteins.
References:
Holliger P., Riechmann L. (1997) A conserved infection pathway for filamentous bacteriophages is suggested by the structure of the membrane penetration domain of the minor coat protein g3p from phage fd. Structure 5(2):265-75.
McCafferty J., Griffiths A.D., Winter G., Chiswell D.J. (1990) Phage antibodies: Filamentous phage displaying antibody variable domains. Nature 348: 552-554
Mead DA, Kemper B (1988) Chimeric single stranded DNA phage-plasmid cloning vectors. Biotchnology 10: 85-102
Smith G.P. (1985) Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virus surface. Science 228: 1315-1317
Stengele I., Bross P., Garcés X., Giray J., Rasched I. (1990) Dissection of functional domains in phage fd adsorption protein. Discrimination between attachment and penetration sites. J Mol Biol. Mar 5;212(1):143-9.
 "
"