Team:ENSPS-Strasbourg/Future
From 2011.igem.org
| (6 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
{{ENSPS-Strasbourg/Temp}} | {{ENSPS-Strasbourg/Temp}} | ||
| - | + | <span id="achiv"></span> | |
= Future plans = | = Future plans = | ||
| - | + | <DIV STYLE="text-align:justify;"> | |
| - | + | ==ACHIEVEMENTS== | |
| - | + | We have proved that electronics and biology can be combined and work together if the synthetic biology field evolve towards more complex model designing. Our software is currently able to simulate the more complex biosystems published, thanks to the powerfulness of electronic tools. But now, we have to extend the concept, and build a new version of the software, more “biologists-friendly”, and more efficient. | |
| - | + | The work we have realized this year is just a part of the software suite, as you can see below : | |
| - | + | ||
| - | <br> | + | [[File:done_scheme.png|border|600px|center|link=]] |
| - | + | ||
| - | + | ||
| - | + | ==NEXT VERSION== | |
| - | + | For our team, the next step will consist of developing this software in two ways. | |
| - | + | The first way consists of improving the software, so as to be as close as possible to the biologists’ needs. Indeed, several things need to be done:<br> | |
| - | + | 1- PARTNERSHIPS WITH BIOLOGISTS: This will go along with partnerships with one or more teams in 2012 to have a better interaction between what we want to and what the biologists really needs. This year is a test for us, and we are open to any suggestion to improve our software. Strong partnerships have to be settled for next year.<br> | |
| - | + | 2- COLLABORATION WITH OTHER SOFTWARE TEAMS: We have a lot of things to learn about other teams, because some teams are close to have realized some functions we want to add in our software suite. We want to have collaborations next year.<br><br><span id="next"></span> | |
| + | The second way is to add several functions to our software, and integrate step by step the missing elements of the suite:<br> | ||
| + | 1- USER INTERFACE : we will integrate a drag&drop interface so as to build the block diagram of the system, which will be a new functionality of the future software.<br> | ||
| + | 2- COMMUNICATION: To integrate a communication between both models is necessary. In the future version, one description will be done for both models, thanks to the graphical user interface.<br> | ||
| + | 3- SIMULATION: The next version will integrate new simulation parameters, so as to increase the possibilities of the system.<br> | ||
| + | |||
| + | </DIV> | ||
Latest revision as of 17:05, 28 October 2011
Future plans
ACHIEVEMENTS
We have proved that electronics and biology can be combined and work together if the synthetic biology field evolve towards more complex model designing. Our software is currently able to simulate the more complex biosystems published, thanks to the powerfulness of electronic tools. But now, we have to extend the concept, and build a new version of the software, more “biologists-friendly”, and more efficient. The work we have realized this year is just a part of the software suite, as you can see below :
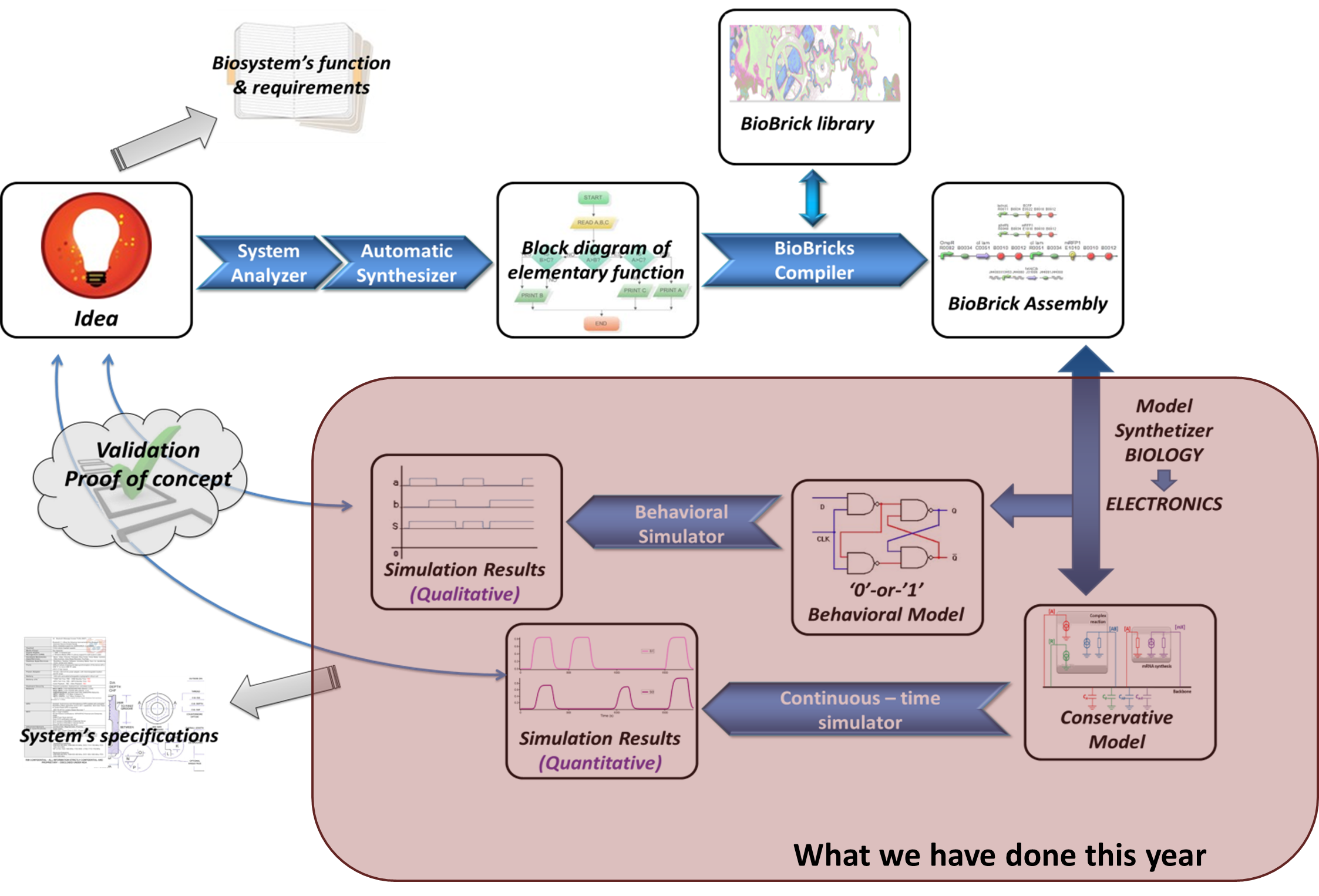
NEXT VERSION
For our team, the next step will consist of developing this software in two ways.
The first way consists of improving the software, so as to be as close as possible to the biologists’ needs. Indeed, several things need to be done:
1- PARTNERSHIPS WITH BIOLOGISTS: This will go along with partnerships with one or more teams in 2012 to have a better interaction between what we want to and what the biologists really needs. This year is a test for us, and we are open to any suggestion to improve our software. Strong partnerships have to be settled for next year.
2- COLLABORATION WITH OTHER SOFTWARE TEAMS: We have a lot of things to learn about other teams, because some teams are close to have realized some functions we want to add in our software suite. We want to have collaborations next year.
The second way is to add several functions to our software, and integrate step by step the missing elements of the suite:
1- USER INTERFACE : we will integrate a drag&drop interface so as to build the block diagram of the system, which will be a new functionality of the future software.
2- COMMUNICATION: To integrate a communication between both models is necessary. In the future version, one description will be done for both models, thanks to the graphical user interface.
3- SIMULATION: The next version will integrate new simulation parameters, so as to increase the possibilities of the system.
 "
"