Team:NTNU Trondheim/Defective Biobricks
From 2011.igem.org
m |
m |
||
| Line 1: | Line 1: | ||
{{:Team:NTNU_Trondheim/NTNU_header}} | {{:Team:NTNU_Trondheim/NTNU_header}} | ||
| + | |||
=Defective biobricks= | =Defective biobricks= | ||
Latest revision as of 08:14, 21 September 2011

Defective biobricks
LacI2 (BBa_K292006)
We found and orderd this part from the parts registry. Since it had RBS and double terminator it would save us from some cloning work.
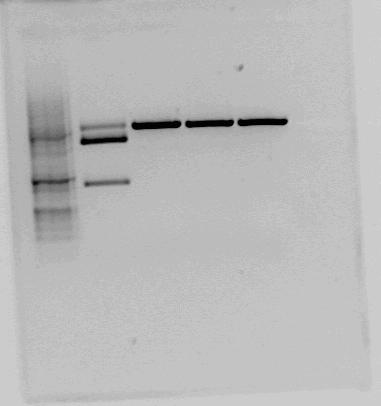
When testcutting the constructs containing BBa_K292006 we got some unexpected results. Therefore we decided to digest the brick with three differnt enzymes with restriction sites in the given biobrick-sequence in addition to EcoR1 and PstI (for looking at the biobrick length) Except from the sample cut with EcorR1 and PstI, the samples were also cut with BglI wich has a recognition site in the pSB1A2 backbone.
Overwiev of restrctions and expected fragmentlengths:
| Restriction enzymes | Expected lengths (bp) |
|---|---|
| EcoR1 + PstI | 2053 (backbone) and 1303 (biobrick) |
| BglI + BclI | 2032 + 1324 |
| BglI + EcoRV | 1760 + 1596 |
| BglI + BanII | 1835 + 1521 |
As we can see on the resulting gel none of the restriction sites inn the LacI2 fragment appear to be present. In additon the LacI2 insert looks smaller than it's supposed to be. This indicates that the biobrick sequence does not match the sequence given in the parts registry.
TetR + p(TetR)+RFP (BBa_K092600) and TetR + p(TetR)+RFP+luxI (BBa_K092700)
At the beginning of the project our idea was to use this TetR regulated system to build the stress sensor. However our experience with the two biobricks is bad. Transformants did not show any signs of fluorescent activity, and the fragment lengths after enzyme digestion were not as expected.
 "
"