Team:NTNU Trondheim/relA
From 2011.igem.org
(→relA) |
(→relA (ppGpp synthase)) |
||
| (11 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
| - | ==relA== | + | ==relA (ppGpp synthase)== |
In a previous study<sup>[1]</sup> the possible fuction of rrnB P1 in a biological containment system was investigated. ppGpp synthase (relA) was over-expressed to investigate the effect on the promoter. They showed that the expression was completely turned off when high amounts of relA was produced<sup>[1]</sup>. relA synthesizes ppGpp when an "empty" amino-acyl t-RNA binds the ribosome. However when it is over-expressed, ppGpp is produced at sufficient levels to inhibit rrnB P1. | In a previous study<sup>[1]</sup> the possible fuction of rrnB P1 in a biological containment system was investigated. ppGpp synthase (relA) was over-expressed to investigate the effect on the promoter. They showed that the expression was completely turned off when high amounts of relA was produced<sup>[1]</sup>. relA synthesizes ppGpp when an "empty" amino-acyl t-RNA binds the ribosome. However when it is over-expressed, ppGpp is produced at sufficient levels to inhibit rrnB P1. | ||
| Line 15: | Line 15: | ||
| - | The gene was amplified from colony PCR, digested and inserted into digested linearized pSB1A3. The sequence of the | + | The gene was amplified from colony PCR, digested and inserted into digested linearized pSB1A3, giving the regular BBa prefix and suffix. The sequence of the BioBrick is given in the text file: |
| - | [[Media: | + | |
| + | [[Media:relABioBrick2.txt| Nucleotide sequence of relA biobrick]] | ||
| + | |||
| + | '''NOTE:''' The gene contains a PstI site at bp 1363-1368. This could be removed with site-specific point mutagenesis, changing CTGCAG → CTACAG or CTGCAA. In our project, we avoided this problem by doing partial digestion in those steps where plasmids containing RelA had to be cut with PstI. | ||
| + | |||
| + | |||
| + | '''Sequencing''' | ||
| + | |||
| + | The BioBrick contains an insert; AAAGAGGAGAAATACTAGAG, immediately before the start-codon. This is probably due to primer-dimerization of som sort during PCR-cloning of the gene. | ||
| + | |||
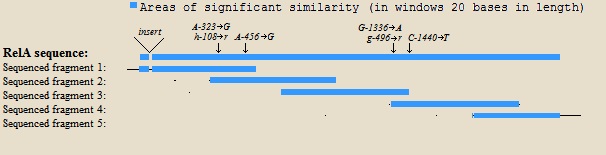
| + | The sequence also contains four nucleotide substitutions compared to the genome of ''E. coli'' K-12. This could be due to mutations in the genome of DH5-alpha or during our cloning. All mutations were supported by two overlapping sequencing fragments. Two of the mutations are synonymous, while the other two give amino-acid changes. The mutations are shown in table 1. An alignement-map of the K12-sequence vs the sequenced fragments is shown in the figure below, with mutations indicated. | ||
| + | |||
| + | |||
| + | Table 1: Mutations in relA BioBrick compared to K-12 sequence relA | ||
| + | {| border=1 | ||
| + | | align="center" |'''DNA mutations''' | ||
| + | | align="center" |'''Amino acid mutations''' | ||
| + | |- | ||
| + | | A 323 --> G||h 108 --> r | ||
| + | |- | ||
| + | | A 456 --> G||none | ||
| + | |- | ||
| + | | G 1336 --> A||g 496 --> r | ||
| + | |- | ||
| + | | C 1440 --> T||none | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | |||
| + | [[File:relA.jpg]] | ||
| - | |||
Latest revision as of 09:38, 21 September 2011

relA (ppGpp synthase)
In a previous study[1] the possible fuction of rrnB P1 in a biological containment system was investigated. ppGpp synthase (relA) was over-expressed to investigate the effect on the promoter. They showed that the expression was completely turned off when high amounts of relA was produced[1]. relA synthesizes ppGpp when an "empty" amino-acyl t-RNA binds the ribosome. However when it is over-expressed, ppGpp is produced at sufficient levels to inhibit rrnB P1.
We wanted to use relA to characterize ppGpp's effect on the rrnB P1 promoter. To use relA in our project, we had to amplify the gene from chromosomal DNA, as it was not found in the Partsregistry. Primers were designed as shown below, giving the full 2234 bp gene plus the [http://openwetware.org/wiki/Synthetic_Biology:BioBricks/Part_fabrication Openwetware prefix and suffix];
relA.fwd: GTTTCTTCGAATTCGCGGCCGCTTCTAGAGATGGTTGCGGTAAGAAGTGCACA
relA.rev: GTTTCTTCCTGCAGCGGCCGCTACTAGTACTAACTCCCGTGCAACCGACG
The gene was amplified from colony PCR, digested and inserted into digested linearized pSB1A3, giving the regular BBa prefix and suffix. The sequence of the BioBrick is given in the text file:
Nucleotide sequence of relA biobrick
NOTE: The gene contains a PstI site at bp 1363-1368. This could be removed with site-specific point mutagenesis, changing CTGCAG → CTACAG or CTGCAA. In our project, we avoided this problem by doing partial digestion in those steps where plasmids containing RelA had to be cut with PstI.
Sequencing
The BioBrick contains an insert; AAAGAGGAGAAATACTAGAG, immediately before the start-codon. This is probably due to primer-dimerization of som sort during PCR-cloning of the gene.
The sequence also contains four nucleotide substitutions compared to the genome of E. coli K-12. This could be due to mutations in the genome of DH5-alpha or during our cloning. All mutations were supported by two overlapping sequencing fragments. Two of the mutations are synonymous, while the other two give amino-acid changes. The mutations are shown in table 1. An alignement-map of the K12-sequence vs the sequenced fragments is shown in the figure below, with mutations indicated.
Table 1: Mutations in relA BioBrick compared to K-12 sequence relA
| DNA mutations | Amino acid mutations |
| A 323 --> G | h 108 --> r |
| A 456 --> G | none |
| G 1336 --> A | g 496 --> r |
| C 1440 --> T | none |
Link to parts registry: [http://partsregistry.org/Part:BBa_K639001 BBa_K639001]
[1] Tedin, K., A. Witte, et al. (1995). "Evaluation of the E. coli ribosomal rrnB P1 promoter and phage-derived lysis genes for the use in a biological containment system: A concept study." Journal of Biotechnology 39(2): 137-148.
 "
"