Team:TU Munich/lab/notebook/part1
From 2011.igem.org
| (27 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
<html> | <html> | ||
| + | <script src="https://2011.igem.org/Team:TU_Munich/slimbox2.js?action=raw&ctype=text/js" type="text/javascript"></script> | ||
| + | <link rel="stylesheet" href="https://2011.igem.org/Team:TU_Munich/slimbox2.css?action=raw&ctype=text/css" type="text/css" media="screen"></link> | ||
| + | |||
<div class="ui-corner-all subcontent tabcontent"> | <div class="ui-corner-all subcontent tabcontent"> | ||
| - | |||
| - | < | + | <h1><span class="mw-headline" id="Light_sensor_systems_and_AND-Gate_cloningPart_I_Wolfgang.2FTobi.2FNico.2FKatharina.2FFlo">Cloning Part I</span></h1> |
| + | <p><b>People: Wolfgang, Tobi, Nico, Katharina, Flo</b></p> | ||
| + | <br> | ||
| + | <h2><span class="mw-headline" id="d05112011">11-05-2011</span></h2> | ||
<h3> <span class="mw-headline">Other Work</span></h3> | <h3> <span class="mw-headline">Other Work</span></h3> | ||
<div class="otherwork"> | <div class="otherwork"> | ||
| - | <p> | + | <p>We request the following parts from the registry:</p> |
<table class="wikitable"> | <table class="wikitable"> | ||
<tr> | <tr> | ||
| Line 30: | Line 35: | ||
</td></tr></table> | </td></tr></table> | ||
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d05122011">12-05-2011</span></h2> |
| - | <p><b>Autoclave broken</b></p><p>Should be working next week again (Andrea is going to tell us | + | <p><b>Autoclave broken</b></p><p>We are not able to prepare any media or sterile materials. Should be working next week again (Andrea is going to tell us).</p><p>-> No agar plate/medium preparation possible right now. </p><p>But all other necessary stuff seems to be available (for electroporation, competent cells, miniprep-kit, DH5alpha strain, BL21 strain).</p> |
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d05192011">19-05-2011</span></h2> |
| - | <p><b>Autoclave working</b> </p> | + | <p><b>Autoclave working</b></p> |
<h3> <span class="mw-headline">Other Work</span></h3> | <h3> <span class="mw-headline">Other Work</span></h3> | ||
<div class="otherwork"> | <div class="otherwork"> | ||
| - | <p> | + | <p>We start preparing Agar-LB Plates with Amp and Kan, as well as normal LB medium.</p> |
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d05202011">20-05-2011</span></h2> |
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
<div class="cloning"> | <div class="cloning"> | ||
<h4> <span class="mw-headline" id="Transformation"> Transformation</span></h4> | <h4> <span class="mw-headline" id="Transformation"> Transformation</span></h4> | ||
<p>Transformation of all red-light sensor parts separately. | <p>Transformation of all red-light sensor parts separately. | ||
| - | + | DH5 alpha cells were transformed using electroporation (V = 1600V) with following constructs:</p><p>Part alias : Part Name - Plasmid - Year of Distribution, Plate Number</p><p>iGEM R1 : R0082 pSB1A3 2010, P1</p><p>iGEM R2 : I732017 pSB1A2 2010, P2</p><p>iGEM R3 : I15010 pSB2K3 2010, P3</p><p>iGEM R4 : K098010 pSB4C5 2010, P3</p><p>Each electroporation was performed with 1.5 µl BioBrick plasmid (each disolved in 10 µl ddH2O before) and 40 µl competent DH5 alpha cells according to the protocol kindly provided by Andrea Mueckl.</p><p>Afterwards cells were incubated in 1 ml SOC medium for 1.5 h at 37 °C, 200 rpm and subsequently plated on LB-agar plates containing antibiotics (50 µl, 100 µl, 200 µl). They were incubated at 37 °C over night and then stored at 4 °C.</p> | |
| - | incubated at | + | |
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d05232011">23-05-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| - | <p>Transformation of all red sensor parts failed</p> | + | <p>Transformation of all red sensor parts failed. This might have been due to a wrong electroporation protocol.</p> |
</div> | </div> | ||
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
<div class="cloning"> | <div class="cloning"> | ||
<h4> <span class="mw-headline" id="Transformation_2">Transformation</span></h4> | <h4> <span class="mw-headline" id="Transformation_2">Transformation</span></h4> | ||
| - | <p>Transformation of one red part sensor (iGEM | + | <p>Transformation of one red part sensor (iGEM R0082) with slightly modified protocol: electroporation was performed at 1500 V with 1 µl Plasmid (part was previously disolved in 10 µl ddH2O)and 40 µl competent DH5 alpha cells. Otherwise same procedure as before.</p> |
</div> | </div> | ||
<h3> <span class="mw-headline">Other Work</span></h3> | <h3> <span class="mw-headline">Other Work</span></h3> | ||
<div class="otherwork"> | <div class="otherwork"> | ||
| - | <p>Making electrocompetent cells according to Andrea's Mueckl protocol</p> | + | <p>Making electrocompetent cells according to Andrea's Mueckl protocol.</p> |
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d05242011">24-05-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| - | <p>Transformation of | + | <p>Transformation of R0082 resulted in 3 colonies.</p> |
</div> | </div> | ||
<h3> <span class="mw-headline">Other Work</span></h3> | <h3> <span class="mw-headline">Other Work</span></h3> | ||
<div class="otherwork"> | <div class="otherwork"> | ||
| - | <p> | + | <p>Inoculation of over-night culture (20 ml) of DH5 alpha cells in Luria-Media for electrocompetent cells.</p><p>Inoculation of over-night culture of one clone picked from the successful second transformation.</p><p>We asked Dr. J. Winter for heatresistant E. coli. We should get them tomorrow after lunch on a plate and can then culture them.</p> |
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d05252011">25-05-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| - | <p> | + | <p>The clone picked yesterday turned out as the right one.</p><p>Dr. J. Winter wants us to come back tomorrow, since bacteria seem to be contaminated</p> |
</div> | </div> | ||
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
<div class="cloning"> | <div class="cloning"> | ||
<h4> <span class="mw-headline" id="MiniPrep">MiniPrep</span></h4> | <h4> <span class="mw-headline" id="MiniPrep">MiniPrep</span></h4> | ||
| - | <p>Testing clone: Mini-Prep using Zymoresearch DNA Kit, | + | <p>Testing of the clone: Mini-Prep using Zymoresearch DNA Kit, with subsequent digestion with EcoRI and PstI in NEB-Buffer 4. Afterwards loading ontol Agarose gel (expected fragments 0.1 kbp (= R0082) and 2,0 kbp (= pSB1A2))</p> |
</div> | </div> | ||
<h3> <span class="mw-headline">Other Work</span></h3> | <h3> <span class="mw-headline">Other Work</span></h3> | ||
<div class="otherwork"> | <div class="otherwork"> | ||
| - | <p>Continue making cells electrocompetent</p> | + | <p>Continue making cells electrocompetent.</p> |
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d05262011">26-05-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| - | <p>Again: J. Winter wants us to come back next day</p> | + | <p>Again: Dr. J. Winter wants us to come back next day, since bacteria seem to be contaminated.</p> |
</div> | </div> | ||
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
| Line 99: | Line 103: | ||
<h4> <span class="mw-headline" id="Transformation_3">Transformation</span></h4> | <h4> <span class="mw-headline" id="Transformation_3">Transformation</span></h4> | ||
<p>Transformation of</p><p>Part alias : Part Name - Plasmid - Year of Distribution, Plate Number | <p>Transformation of</p><p>Part alias : Part Name - Plasmid - Year of Distribution, Plate Number | ||
| - | </p><p>iGEM R2 : I732017 pSB1A2 2010, P2</p><p>iGEM R3 : I15010 pSB2K3 2010, P3</p><p>iGEM R4 : K098010 pSB4C5 2010, P3 </p><p>using | + | </p><p>iGEM R2 : I732017 pSB1A2 2010, P2</p><p>iGEM R3 : I15010 pSB2K3 2010, P3</p><p>iGEM R4 : K098010 pSB4C5 2010, P3 </p><p>We were using 1510 V and 1 µl DNA for transformation. The DNA was just added to the cells and gently mixed by stirring it with the pipette tip. The protocol used was the one from Andrea Meyer. Of the 1 ml SOC culture, 50 µl were spread out directly on Agar plates. Rest was spun down (2000 rpm, 1 min) and pellet was resuspended in 100 to 150 µl. Subsequently, 50 µl were spread out.</p> |
| - | Of the | + | |
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d05272011">27-05-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| - | <p>successful clones from transformed cells with iGEM | + | <p>There are successful clones from transformed cells with iGEM I732017 and iGEM K098010, but no clones from iGEM I15010 at all</p><p>Again: Dr. J. Winter wants us to come back next week, as there is still contamination in the culture.</p> |
</div> | </div> | ||
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
| Line 112: | Line 115: | ||
<h4> <span class="mw-headline" id="Transformation_4">Transformation</span></h4> | <h4> <span class="mw-headline" id="Transformation_4">Transformation</span></h4> | ||
<p>Transformation of:</p><p>Part alias : Part Name - Plasmid - Year of Distribution, Plate Number | <p>Transformation of:</p><p>Part alias : Part Name - Plasmid - Year of Distribution, Plate Number | ||
| - | </p><p>iGEM R3 : I15010 pSB2K3 2011, P3</p><p>iGEM tRNA : K228001, pSB1A2 2011, P4</p><p>iGEM Term : B0015 pSB1AK3 2011, P1</p><p>iGEM RBS : J44001 pSB1A2 2011, P1</p><p>iGEM T7 : I712074 pSB1AK8 2011, P1</p><p> | + | </p><p>iGEM R3 : I15010 pSB2K3 2011, P3</p><p>iGEM tRNA : K228001, pSB1A2 2011, P4</p><p>iGEM Term : B0015 pSB1AK3 2011, P1</p><p>iGEM RBS : J44001 pSB1A2 2011, P1</p><p>iGEM T7 : I712074 pSB1AK8 2011, P1</p><p>Since the transformation with iGEM I15010 did not work, we increased the amount of plasmid to 2 µl. Afterwards the procedure was performed as mentioned above and the cells were incubated over night (15 h) at 37 °C and cooled until monday at 4 °C.</p> |
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d05302011">30-05-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| - | <p> | + | <p>We have successful clones from transformed cells with iGEM K228001 (tRNA), iGEM B0015 (Term), iGEM J44001 (RBS), iGEM I712074 (T7).</p><p>Finally, we received BM28 cells from Dr. J. Winter (there is an existing paper in JBC from her about the bacteria).</p> |
</div> | </div> | ||
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
<div class="cloning"> | <div class="cloning"> | ||
<h4> <span class="mw-headline" id="Transformation_5">Transformation</span></h4> | <h4> <span class="mw-headline" id="Transformation_5">Transformation</span></h4> | ||
| - | <p>Transformation of | + | <p>Transformation of I15010 once more using BL21 and inducing pSB2K3 plasmid using 1 mM IPTG in SOC-Medium.</p> |
</div> | </div> | ||
<h3> <span class="mw-headline">Other Work</span></h3> | <h3> <span class="mw-headline">Other Work</span></h3> | ||
<div class="otherwork"> | <div class="otherwork"> | ||
| - | <p> | + | <p>Inoculation of over-night cultures of clones from iGEM K228001 (tRNA), iGEM B0015 (Term), iGEM J44001 (RBS), iGEM I712074 (T7), iGEM I732017 und iGEM K098010</p> |
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d05312011">31-05-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| - | <p> | + | <p>The transformation of iGEM I15010 (R3) did not work.</p><p>We measured plasmid concentrations from MiniPreps (see below) using nano-drop:</p><p>- iGEM I712074 (T7): 62 ng/µl</p><p>- iGEM K098010 (R4): 82,5 ng/µl </p><p>- iGEM K228001 (tRNA): 98 ng/µl</p><p>- iGEM I732017 (R2): 94 ng/µl</p><p>- iGEM B0015 (Term): 141 ng/µl</p><p>- iGEM J44001 (RBS): 108 ng/µl</p><p>- iGEM R0082 (R1): 59 ng/µl</p> |
</div> | </div> | ||
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
<div class="cloning"> | <div class="cloning"> | ||
<h4> <span class="mw-headline" id="MiniPrep_2">MiniPrep</span></h4> | <h4> <span class="mw-headline" id="MiniPrep_2">MiniPrep</span></h4> | ||
| - | <p>Plasmid Isolation (MiniPrep) from all over night cultures</p> | + | <p>Plasmid Isolation (MiniPrep) from all over night cultures was conducted and the concentrations were measured.</p> |
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d06012011">01-06-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| - | <p> | + | <p>A SDS-PAGE was conducted from the cultures induced with IPTG on 50-05-2011. The cells were boiled for 5 min at 95 °C in loading buffer. The gel ran inconclusive, next time there should be alonger run with higher voltage. Only 1/3 of the lane was used by the peptides. It had a very bad resolution. However, the dying step worked properly.</p> |
</div> | </div> | ||
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
<div class="cloning"> | <div class="cloning"> | ||
<h4> <span class="mw-headline" id="Digestion">Digestion</span></h4> | <h4> <span class="mw-headline" id="Digestion">Digestion</span></h4> | ||
| - | <p>- | + | <p>- A restriction digestion of the parts gained by MiniPrep was conducted using EcoRI and PstI in NEB-Buffer 4. Afterwards, they were applied onto an agarose gel.</p><p>- Properties of the gel run with the digested samples: 100 V, 1 h 10 Min, 1 % agarose.</p><p>- SYBR Gold staining: 2 µl on shaking plate for 30 mins at room temperature (RT).</p> |
<h4> <span class="mw-headline" id="Transformation_6">Transformation</span></h4> | <h4> <span class="mw-headline" id="Transformation_6">Transformation</span></h4> | ||
| - | <p>Transformation of | + | <p>Transformation of DH5 alpha with:</p><p>iGEM I15010 (R3) (see above) on ampicilin (A, Amp), chloramphenicol (C, Cm) and kanamycin (K, Kan) containing resistance plates;</p><p>iGEM High Copy Plasmid, Plate 1 2011 1G, psB1A3 with insert: BBa_J04450, Resistance: A;</p><p>iGEM Low Copy Plasmid, Plate 1 2011 3C, psB3C5 with insert: BBa_J04450, Resistance: C.</p><p>They were plated on agar plates carrying the appropriate antibiotic resistances and incubated at 37 °C over night.</p> |
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d06032011">03-06-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| - | <p> | + | <p>Another SDS-PAGE gel run again inconclusive. Furthermore during the run, there was a loss of electrical power because of some blackout in the building.</p><p>Transformed cells grew very well over night, with a few clones of iGEM I15010 (R3) on kanamycin resistance plate found. This is evidence for a contamination, because the incubator did not cool down to 4 °C over thursday.</p><p>The transformed plasmid-cells grew also very good - now featured in red.</p> |
</div> | </div> | ||
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
<div class="cloning"> | <div class="cloning"> | ||
<h4> <span class="mw-headline" id="Digestion_2">Digestion</span></h4> | <h4> <span class="mw-headline" id="Digestion_2">Digestion</span></h4> | ||
| - | <p> | + | <p>An agarose gel was run with digestions from yesterday, using the following settings: 90 V, 1 h 30 Min, 1 % agarose. |
| - | </p><p> | + | </p><p>SYBR Gold staining: 2 µl on shaking plate for 30 mins at RT.</p> |
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d06062011">06-06-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| - | <p> | + | <p>The agarose gel was again inconclusive. There may be too less material, since no bands around 100 bp could be detected at all.</p> |
</div> | </div> | ||
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
<div class="cloning"> | <div class="cloning"> | ||
<h4> <span class="mw-headline" id="Digestion_3">Digestion</span></h4> | <h4> <span class="mw-headline" id="Digestion_3">Digestion</span></h4> | ||
| - | <p> | + | <p>Another digestion was conducted and analyzed on an agaorse gel. It ran with newly digested and undigested plasmid as control: 100 V, 1 h 30 Min, 1 % agarose.</p> |
<h4> <span class="mw-headline" id="Transformation_7">Transformation</span></h4> | <h4> <span class="mw-headline" id="Transformation_7">Transformation</span></h4> | ||
| - | <p>Transformation of R3 in E. coli D1210</p> | + | <p>Transformation of I15010 (R3) in E. coli D1210</p> |
</div> | </div> | ||
<h3> <span class="mw-headline">Other Work</span></h3> | <h3> <span class="mw-headline">Other Work</span></h3> | ||
<div class="otherwork"> | <div class="otherwork"> | ||
| - | <p>Over-night culture of: </p><p>iGEM High Copy Plasmid, Plate 1 2011 1G, psB1A3 with insert: BBa_J04450, Resistance: A</p><p>iGEM Low Copy Plasmid, Plate 1 2011 3C, psB3C5 with insert: BBa_J04450, Resistance: C</p> | + | <p>Over-night culture of the following parts were inoculated: </p><p>iGEM High Copy Plasmid, Plate 1 2011 1G, psB1A3 with insert: BBa_J04450, Resistance: A;</p><p>iGEM Low Copy Plasmid, Plate 1 2011 3C, psB3C5 with insert: BBa_J04450, Resistance: C.</p> |
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d06072011">07-06-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| - | <p> | + | <p>The transformation of I15010 (R3) finally worked using E. coli D1210 cells.</p><p>MiniPrep of the over night culture of iGEM Low Copy Plasmid, Plate 1 2011 3C, psB3C5 with insert: BBa_J04450, Resistance: C resulted in 101 ng/µl in a total volume of 100 µl.</p><p>We have run out of DNA-Plasmid Preparation Kit. No more preparations are possible during the next days, until we have a new kit! </p><p>A new digestion was conducted. The subsequent gel still was inconclusive.</p> |
</div> | </div> | ||
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
<div class="cloning"> | <div class="cloning"> | ||
<h4> <span class="mw-headline" id="Digestion_4">Digestion</span></h4> | <h4> <span class="mw-headline" id="Digestion_4">Digestion</span></h4> | ||
| - | <p>2 % Agarose with | + | <p>The digestion was repeated. In order to have a more sensitive detection of DNA on the gel, we now use ethidium bromide for staining of the gel. The following settings are used: 2 % Agarose with ethidium bromide, 90 V, 1.5 h.</p><p>Digestion of 2 µg Plasmid DNA with high fidelity 1.: EcoRI and PstI enzymes and 2.: just with PstI was conducted.</p><p><br /></p> |
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d06082011">08-06-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| Line 205: | Line 208: | ||
</p><p><br /></p> | </p><p><br /></p> | ||
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d06092011">09-06-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| Line 226: | Line 229: | ||
<p>10% PAGE of RBS and T7 </p><p><br /></p> | <p>10% PAGE of RBS and T7 </p><p><br /></p> | ||
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d06102011">10-06-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| Line 239: | Line 242: | ||
</p><p><br /></p> | </p><p><br /></p> | ||
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d06142011">14-06-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| Line 252: | Line 255: | ||
(Cut R4, R2)</p> | (Cut R4, R2)</p> | ||
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d06162011">16-06-2011</span></h2> |
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
<div class="cloning"> | <div class="cloning"> | ||
<p>Cloning of the red light sensor with parts R1,R2,R4 (see cloning draft)</p><p>ONC of part containing colonies in 5ml liquid culture</p><p><br /></p> | <p>Cloning of the red light sensor with parts R1,R2,R4 (see cloning draft)</p><p>ONC of part containing colonies in 5ml liquid culture</p><p><br /></p> | ||
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d06172011">17-06-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| Line 273: | Line 276: | ||
<p>MiniPreps of Parts</p><p><br /></p> | <p>MiniPreps of Parts</p><p><br /></p> | ||
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d06202011">20-06-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| Line 287: | Line 290: | ||
<p>-Backup Plates of existing clones</p><p><br /></p> | <p>-Backup Plates of existing clones</p><p><br /></p> | ||
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d06212011">21-06-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| Line 301: | Line 304: | ||
<p>-Cloning of the blue sensor (only step1) with parts K228000, RBS (see cloning draft)</p><p><br /></p> | <p>-Cloning of the blue sensor (only step1) with parts K228000, RBS (see cloning draft)</p><p><br /></p> | ||
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d06222011">22-06-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| Line 314: | Line 317: | ||
lanes: 2log ladder, RBS undigested (=ud), RBS digested (dig), K22800 ud, K22800 dig, A1 ud, A1 dig, Term ud, Term dig, L1 ud, L1 dig, L2 ud, L2 dig, R2 ud, R2 dig | lanes: 2log ladder, RBS undigested (=ud), RBS digested (dig), K22800 ud, K22800 dig, A1 ud, A1 dig, Term ud, Term dig, L1 ud, L1 dig, L2 ud, L2 dig, R2 ud, R2 dig | ||
6 µl each lane | 6 µl each lane | ||
| - | gel: <a href="/wiki/ | + | gel: <a href="/wiki/images/b/b8/220611_digest_control_2.jpg" class="image" rel="lightbox"><img alt="220611 digest control 2" src="/wiki/images/b/b8/220611_digest_control_2.jpg" width="500" height="372" /></a></p> |
<h4> <span class="mw-headline" id="Transformation_8">Transformation</span></h4> | <h4> <span class="mw-headline" id="Transformation_8">Transformation</span></h4> | ||
<p>-Transformation of both ligations (red and blue light) and incubation on plates over night:</p><p>- blue light on amp and kan</p><p>- red light I on amp plate not protected from light, lightning during electroporation</p><p>- red light II on amp 1 plate protected from light, 1 plate not protected from light</p> | <p>-Transformation of both ligations (red and blue light) and incubation on plates over night:</p><p>- blue light on amp and kan</p><p>- red light I on amp plate not protected from light, lightning during electroporation</p><p>- red light II on amp 1 plate protected from light, 1 plate not protected from light</p> | ||
| Line 332: | Line 335: | ||
low copy plasmid 66.5</p><p><br /></p> | low copy plasmid 66.5</p><p><br /></p> | ||
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d06242011">24-06-2011</span></h2> |
<h3> <span class="mw-headline">Other Work</span></h3> | <h3> <span class="mw-headline">Other Work</span></h3> | ||
<div class="otherwork"> | <div class="otherwork"> | ||
<p>New agarose plates</p><p><br /></p> | <p>New agarose plates</p><p><br /></p> | ||
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d06272011">27-06-2011</span></h2> |
<h3> <span class="mw-headline">Other Work</span></h3> | <h3> <span class="mw-headline">Other Work</span></h3> | ||
<div class="otherwork"> | <div class="otherwork"> | ||
<p>Design of new cloning strategy together with instructors</p><p><br /></p> | <p>Design of new cloning strategy together with instructors</p><p><br /></p> | ||
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d06282011">28-06-2011</span></h2> |
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
<div class="cloning"> | <div class="cloning"> | ||
| Line 351: | Line 354: | ||
</p> | </p> | ||
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d06292011">29-06-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| Line 361: | Line 364: | ||
<p>Digestion (EcoRIxPstI) of K238013, tRNA in pSB3C5, blue light construct (j44001-K22800-B0015), Reporter construct (T7-R2)</p><p><br /></p> | <p>Digestion (EcoRIxPstI) of K238013, tRNA in pSB3C5, blue light construct (j44001-K22800-B0015), Reporter construct (T7-R2)</p><p><br /></p> | ||
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d06302011">30-06-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| Line 374: | Line 377: | ||
</p> | </p> | ||
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d07012011">01-07-2011</span></h2> |
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
<div class="cloning"> | <div class="cloning"> | ||
| Line 398: | Line 401: | ||
Making of electrocompetent cells of DH5alpha and BM28</p><p><br /></p> | Making of electrocompetent cells of DH5alpha and BM28</p><p><br /></p> | ||
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d07042011">04-07-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| - | <p><a href="/wiki/ | + | <p><a href="/wiki/images/f/fc/040711_Plasmid2_analyse_inverse.jpg" class="image" rel="lightbox"><img alt="040711 Plasmid2 analyse inverse" src="/wiki/images/f/fc/040711_Plasmid2_analyse_inverse.jpg" width="400" height="298" /></a></p><p><br /> |
</p> | </p> | ||
</div> | </div> | ||
| Line 409: | Line 412: | ||
<h4> <span class="mw-headline" id="Agarose">Agarose</span></h4> | <h4> <span class="mw-headline" id="Agarose">Agarose</span></h4> | ||
<p>Agarose Gel of ladder, R2, T7, Pol, lac-Pol(R2-Pol) ligation, T7-R2 ligation</p><p>analytical agarose gelelectrophoresis of the PCR samples from 01-07-11: (1%, 1x TBE, 120 V, 400 mA, 90 min)</p><p>10 µl PCR sample + 2 µl loading buffer -> 10 µl per well | <p>Agarose Gel of ladder, R2, T7, Pol, lac-Pol(R2-Pol) ligation, T7-R2 ligation</p><p>analytical agarose gelelectrophoresis of the PCR samples from 01-07-11: (1%, 1x TBE, 120 V, 400 mA, 90 min)</p><p>10 µl PCR sample + 2 µl loading buffer -> 10 µl per well | ||
| - | </p><p>lanes:</p><p>1.) 2-log</p><p>2) negative control: primer (10 µM) + water</p><p>3) DNA: 1,5 ng; primer (10 µM)</p><p>4) DNA: 15 ng; primer (10 µM)</p><p>5) DNA: 1,5 ng; primer (5 µM)</p><p>6) DNA: 15 ng; primer (5 µM)</p><p>7) DNA: 1,5 ng; primer (1 µM)</p><p>8) DNA: 15 ng; primer (1 µM)</p><p>9) 2-log</p><p><a href="/wiki/ | + | </p><p>lanes:</p><p>1.) 2-log</p><p>2) negative control: primer (10 µM) + water</p><p>3) DNA: 1,5 ng; primer (10 µM)</p><p>4) DNA: 15 ng; primer (10 µM)</p><p>5) DNA: 1,5 ng; primer (5 µM)</p><p>6) DNA: 15 ng; primer (5 µM)</p><p>7) DNA: 1,5 ng; primer (1 µM)</p><p>8) DNA: 15 ng; primer (1 µM)</p><p>9) 2-log</p><p><a href="/wiki/images/d/db/040711-PCR-redlight-1.jpg" class="image" rel="lightbox"><img alt="040711-PCR-redlight-1" src="/wiki/images/d/db/040711-PCR-redlight-1.jpg" width="300" height="281" /></a></p><p><br /></p> |
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d07052011">05-07-2011</span></h2> |
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
<div class="cloning"> | <div class="cloning"> | ||
| Line 422: | Line 425: | ||
Extension: 72°C, 2´ 30 s | Extension: 72°C, 2´ 30 s | ||
</pre> | </pre> | ||
| - | <p>3) Final extension: 72°C, 8'</p><p>4) Hold: 4°C, oo</p><p><b>Samples</b></p><p>PCR-samples were prepared with HF- and GC-buffer, respectively. End volume of the reaction setups was 50 ul.</p><p>DMSO-concentration was 100%; primer concentration was 10 uM, which equals an end conc. of 500 nM (2.5ul) or 200 nM (1ul).</p><p>DNA-template [150 ng/ul] was diluted 1:100 to a conc. of 1.5 ng/ul -> 1 ul of dilution was used per reaction.</p><p>Samples were prepared as follows (same setup for HF- and GC-buffer):</p><p><a href="/wiki/ | + | <p>3) Final extension: 72°C, 8'</p><p>4) Hold: 4°C, oo</p><p><b>Samples</b></p><p>PCR-samples were prepared with HF- and GC-buffer, respectively. End volume of the reaction setups was 50 ul.</p><p>DMSO-concentration was 100%; primer concentration was 10 uM, which equals an end conc. of 500 nM (2.5ul) or 200 nM (1ul).</p><p>DNA-template [150 ng/ul] was diluted 1:100 to a conc. of 1.5 ng/ul -> 1 ul of dilution was used per reaction.</p><p>Samples were prepared as follows (same setup for HF- and GC-buffer):</p><p><a href="/wiki/images/6/61/060711-PCR-Ansatz.jpg" class="image" rel="lightbox"><img alt="060711-PCR-Ansatz" src="/wiki/images/6/61/060711-PCR-Ansatz.jpg" width="800" height="217" /></a> |
</p> | </p> | ||
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d07062011">06-07-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| - | <p><a href="/wiki/ | + | <p><a href="/wiki/images/2/23/Gel_Verdau_RBS-K228000.jpg" class="image" rel="lightbox"><img alt="Gel Verdau RBS-K228000" src="/wiki/images/2/23/Gel_Verdau_RBS-K228000.jpg" width="400" height="388" /></a></p><p><br /> |
<b>HF-buffer samples gel analysis:</b> | <b>HF-buffer samples gel analysis:</b> | ||
| - | </p><p><a href="/wiki/ | + | </p><p><a href="/wiki/images/8/85/060711-PCR-Testgel-HF-invers-beschriftet.jpg" class="image" rel="lightbox"><img alt="060711-PCR-Testgel-HF-invers-beschriftet" src="/wiki/images/8/85/060711-PCR-Testgel-HF-invers-beschriftet.jpg" width="300" height="461" /></a> -> PCR seems to have been successful, amplicon is at about 4kb!</p><p><b>QC-buffer samples gel analysis (before and after cut-out):</b></p><p><br /> |
| - | <a href="/wiki/ | + | <a href="/wiki/images/c/cb/QC-buffer-samples-PCR-vorher%2Bnachher.jpg" class="image" rel="lightbox"><img alt="QC-buffer-samples-PCR-vorher+nachher" src="/wiki/images/c/cb/QC-buffer-samples-PCR-vorher%2Bnachher.jpg" width="800" height="407" /></a> -> PCR also successful, cut-out of 4 bands, DNA prep and pooling samples</p> |
</div> | </div> | ||
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
| Line 438: | Line 441: | ||
<h4> <span class="mw-headline" id="Digestion_8">Digestion</span></h4> | <h4> <span class="mw-headline" id="Digestion_8">Digestion</span></h4> | ||
<p>Digestion of J44001, K228000</p><p>Preparative gel run with the digested parts (1% agarose, 1xTAE, 120 V 400 mA, 1.5 h)</p><p>Isolation of the bands out of the gel</p><p>Preparation with a gel isolation kit (freeze ´n squeeze, bio-rad)</p><p>work-up of PCR-run (05-07-2011): gel analysis (1% agarose, 1xTBE, 120 V, 400 mA, 1.5 h) of HF-buffer-samples</p><p>gel analysis (1% agarose, 1xTBE, 120 V, 400 mA, 1.5 h) of GC-buffer-samples</p><p>isolation of right bands out of the GC-gel</p><p>DNA preparation with a gel isolation kit (freeze ´n squeeze, bio-rad)</p><p>restriction digest with EcoRI and SpeI</p> | <p>Digestion of J44001, K228000</p><p>Preparative gel run with the digested parts (1% agarose, 1xTAE, 120 V 400 mA, 1.5 h)</p><p>Isolation of the bands out of the gel</p><p>Preparation with a gel isolation kit (freeze ´n squeeze, bio-rad)</p><p>work-up of PCR-run (05-07-2011): gel analysis (1% agarose, 1xTBE, 120 V, 400 mA, 1.5 h) of HF-buffer-samples</p><p>gel analysis (1% agarose, 1xTBE, 120 V, 400 mA, 1.5 h) of GC-buffer-samples</p><p>isolation of right bands out of the GC-gel</p><p>DNA preparation with a gel isolation kit (freeze ´n squeeze, bio-rad)</p><p>restriction digest with EcoRI and SpeI</p> | ||
| - | < | + | <h4> <span class="mw-headline">MiniPrep</span></h4> |
| - | + | ||
| - | + | ||
<p>mini-prep of culture (4 ml) from DH5alpha (05-07-2011) (qiagen qiaprep spin miniprep kit) -> 82 ng/µl in 50 µl elution buffer</p><p>restriction digest of 0.5 µg DNA with EcoRI and PstI</p><p>gel analysis of restriction digest (1% agarose, 1xTAE, 120 V, 400 mA, 1.5 h)</p><p><br /></p> | <p>mini-prep of culture (4 ml) from DH5alpha (05-07-2011) (qiagen qiaprep spin miniprep kit) -> 82 ng/µl in 50 µl elution buffer</p><p>restriction digest of 0.5 µg DNA with EcoRI and PstI</p><p>gel analysis of restriction digest (1% agarose, 1xTAE, 120 V, 400 mA, 1.5 h)</p><p><br /></p> | ||
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d07072011">07-07-2011</span></h2> |
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
<div class="cloning"> | <div class="cloning"> | ||
| Line 450: | Line 451: | ||
<p>Ligation (total volume: 50 µl; 1.5 µg of each DNA sample; 5 µl T4-ligase buffer (10x); 1 µl Quick-Ligase; add to 50 µl with nf H20; 60`@ 37°C)</p><p>Ligation of:</p><p>1. K228000 + RBS</p><p>2. K322127 + supD-tRNA</p><p>3. lacZ + T7-Promotor</p> | <p>Ligation (total volume: 50 µl; 1.5 µg of each DNA sample; 5 µl T4-ligase buffer (10x); 1 µl Quick-Ligase; add to 50 µl with nf H20; 60`@ 37°C)</p><p>Ligation of:</p><p>1. K228000 + RBS</p><p>2. K322127 + supD-tRNA</p><p>3. lacZ + T7-Promotor</p> | ||
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d07082011">08-07-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| - | <p>Ligations failed</p><p><a href="/wiki/ | + | <p>Ligations failed</p><p><a href="/wiki/images/a/a2/080711_ligationen_failed.jpg" class="image" rel="lightbox"><img alt="080711 ligationen failed" src="/wiki/images/a/a2/080711_ligationen_failed.jpg" width="500" height="506" /></a></p><p><br /></p> |
</div> | </div> | ||
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
| Line 461: | Line 462: | ||
</p><p>30 µl DNA + 6 µl loading dye (6x) -> 36 µl, 35 µl loaded on gel</p><p>7,5 µl 2-log DNA ladder</p><p><br /></p> | </p><p>30 µl DNA + 6 µl loading dye (6x) -> 36 µl, 35 µl loaded on gel</p><p>7,5 µl 2-log DNA ladder</p><p><br /></p> | ||
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d07112011">11-07-2011</span></h2> |
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
<div class="cloning"> | <div class="cloning"> | ||
| Line 470: | Line 471: | ||
<h4> <span class="mw-headline" id="Restriction">Restriction</span></h4> | <h4> <span class="mw-headline" id="Restriction">Restriction</span></h4> | ||
<p><b>Restriction digest</b> (total volume: 50 µl; Enzymes: 1 µl each; 5 µl NEB 4 buffer (10x); incubation: 1 h @ 37°C; inactivation: 20 ' @ 80 °C) of PCR samples 2 and 4 (HF-buffer) from 05-07-11:</p><p>DNA: 10 µl (sample 2: 1 µl primer [10 µM], -DMSO; sample 4: 1 µl primer [10 µM], +DMSO)</p><p>H20: 33 µl</p><p>Enzymes: E, S</p><p><br /> | <p><b>Restriction digest</b> (total volume: 50 µl; Enzymes: 1 µl each; 5 µl NEB 4 buffer (10x); incubation: 1 h @ 37°C; inactivation: 20 ' @ 80 °C) of PCR samples 2 and 4 (HF-buffer) from 05-07-11:</p><p>DNA: 10 µl (sample 2: 1 µl primer [10 µM], -DMSO; sample 4: 1 µl primer [10 µM], +DMSO)</p><p>H20: 33 µl</p><p>Enzymes: E, S</p><p><br /> | ||
| - | <b>Preparative agarosegel</b> (1%, 1xTAE, 8 wells, 120 V, 400 mA, 90'):</p><p>35 µl restriction digest + 7 µl loading dye (6x) -> 42 µl; 40 µl loaded on gel</p><p>7 µl 2-log DNA ladder</p><p>lanes:</p><p>1.) 2-log</p><p>2.) PCR-sample 2 from 05-07-11</p><p>3.) PCR-sample 4 from 05-07-11</p><p><a href="/ | + | <b>Preparative agarosegel</b> (1%, 1xTAE, 8 wells, 120 V, 400 mA, 90'):</p><p>35 µl restriction digest + 7 µl loading dye (6x) -> 42 µl; 40 µl loaded on gel</p><p>7 µl 2-log DNA ladder</p><p>lanes:</p><p>1.) 2-log</p><p>2.) PCR-sample 2 from 05-07-11</p><p>3.) PCR-sample 4 from 05-07-11</p><p><a href="https://static.igem.org/mediawiki/2011/3/3f/11711_pcr_pr%C3%A4p.jpg" class="image" rel="lightbox"><img alt="11711 pcr präp" src="https://static.igem.org/mediawiki/2011/3/3f/11711_pcr_pr%C3%A4p.jpg" width="250" height="433" /></a> <a href="/wiki/images/e/e8/11711_pcr_pr%C3%A4p_danach.jpg" class="image" rel="lightbox"><img alt="11711 pcr präp danach" src="/wiki/images/e/e8/11711_pcr_pr%C3%A4p_danach.jpg" width="240" height="429" /></a></p> |
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d07122011">12-07-2011</span></h2> |
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
<div class="cloning"> | <div class="cloning"> | ||
| Line 480: | Line 481: | ||
</p><p>- SupD-tRNA: 150 ng/µl -> 10 µl; H20: 33 µl; Enzymes: E, X (#1, #2: double preparation for validation of different subsequent DNA purification steps)</p><p>- K238013 (blue light promotor): 233 ng/µl -> 6,5 µl; H20: 36,5 µl; Enzymes: E, X</p><p>- B0015 (Term.): 141 ng/µl -> 10,5 µl; H20: 32,5 µl; Enzymes: E, S</p><p>- RBS: 108 ng/µl -> 14 µl; H20: 29 µl; Enzymes: S, P</p><p><br /> | </p><p>- SupD-tRNA: 150 ng/µl -> 10 µl; H20: 33 µl; Enzymes: E, X (#1, #2: double preparation for validation of different subsequent DNA purification steps)</p><p>- K238013 (blue light promotor): 233 ng/µl -> 6,5 µl; H20: 36,5 µl; Enzymes: E, X</p><p>- B0015 (Term.): 141 ng/µl -> 10,5 µl; H20: 32,5 µl; Enzymes: E, S</p><p>- RBS: 108 ng/µl -> 14 µl; H20: 29 µl; Enzymes: S, P</p><p><br /> | ||
preparative agarosegel (1%, 1xTAE, 8 wells, 120 V, 400 mA, 90'):</p><p>35 µl restriction digest + 7 µl loading dye (6x) -> 42 µl; 40 µl loaded on gel | preparative agarosegel (1%, 1xTAE, 8 wells, 120 V, 400 mA, 90'):</p><p>35 µl restriction digest + 7 µl loading dye (6x) -> 42 µl; 40 µl loaded on gel | ||
| - | </p><p>7 µl 2-log DNA ladder</p><p>1) DNA ladder</p><p>2) lacZ</p><p>3) T7 Promotor</p><p>4) supD tRNA</p><p>5) K328013</p><p>6) RBS</p><p><a href="/ | + | </p><p>7 µl 2-log DNA ladder</p><p>1) DNA ladder</p><p>2) lacZ</p><p>3) T7 Promotor</p><p>4) supD tRNA</p><p>5) K328013</p><p>6) RBS</p><p><a href="https://static.igem.org/mediawiki/2011/4/4d/120711_verdau_1_prozent_gel-nachher.jpg" class="image" rel="lightbox"><img alt="120711 verdau 1 prozent gel-nachher" src="https://static.igem.org/mediawiki/2011/4/4d/120711_verdau_1_prozent_gel-nachher.jpg" width="300" height="269" /></a></p><p><br /> |
preparative agarosegel (2%, 1xTAE, 8 wells, 120 V, 400 mA, 90'):</p><p>35 µl restriction digest + 7 µl loading dye (6x) -> 42 µl; 40 µl loaded on gel</p><p>7 µl 2-log DNA ladder</p><p>0) DNA ladder (pipette tip fell off)</p><p>1) DNA ladder</p><p>2) B0015 (Term.) | preparative agarosegel (2%, 1xTAE, 8 wells, 120 V, 400 mA, 90'):</p><p>35 µl restriction digest + 7 µl loading dye (6x) -> 42 µl; 40 µl loaded on gel</p><p>7 µl 2-log DNA ladder</p><p>0) DNA ladder (pipette tip fell off)</p><p>1) DNA ladder</p><p>2) B0015 (Term.) | ||
| - | </p><p><a href="/ | + | </p><p><a href="https://static.igem.org/mediawiki/2011/a/a8/120711_b0015_2_prozent_gel.jpg" class="image" rel="lightbox"><img alt="120711 b0015 2 prozent gel" src="https://static.igem.org/mediawiki/2011/a/a8/120711_b0015_2_prozent_gel.jpg" width="260" height="460" /></a> <a href="/wiki/images/3/39/120711_b0015_2_prozent_gel-nachher.jpg" class="image" rel="lightbox"><img alt="120711 b0015 2 prozent gel-nachher" src="/wiki/images/3/39/120711_b0015_2_prozent_gel-nachher.jpg" width="250" height="458" /></a></p> |
<h4> <span class="mw-headline" id="Ligation_3">Ligation</span></h4> | <h4> <span class="mw-headline" id="Ligation_3">Ligation</span></h4> | ||
<p>Ligation (total volume: 30 µl; 10 µl of each DNA sample; 3 µl T4-ligase buffer (10x); 1 µl Quick-Ligase; 6 µl nf H20; 30`@ 37°C)</p><p>Ligation of: </p><p>1. lacZ + T7-Promotor</p><p>2. K322127 (PCR-Product) + supD-tRNA</p><p>3. K238013 + B0015</p> | <p>Ligation (total volume: 30 µl; 10 µl of each DNA sample; 3 µl T4-ligase buffer (10x); 1 µl Quick-Ligase; 6 µl nf H20; 30`@ 37°C)</p><p>Ligation of: </p><p>1. lacZ + T7-Promotor</p><p>2. K322127 (PCR-Product) + supD-tRNA</p><p>3. K238013 + B0015</p> | ||
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d07132011">13-07-2011</span></h2> |
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
<div class="cloning"> | <div class="cloning"> | ||
| Line 500: | Line 501: | ||
<div class="otherwork"> | <div class="otherwork"> | ||
<p>Analytical agarosegel [130711-ligation-control.tif] (1%, 1xTBE, 20 wells, 120 V, 400 mA, 90') for control of ligations from 12-07-11:</p><p>10 µl ligation sample + 2 µl loading dye (6x) -> 12 µl, 10 µl loaded on gel</p><p>5 µl 2-log DNA ladder</p><p>lanes:</p><p>1) 2-log</p><p>2) lacZ + T7-promotor AFTER purification</p><p>3) lacZ + T7-promotor BEFORE purification</p><p>4) PCR-product (from K322127) + supD-tRNA AFTER purification</p><p>5) PCR-product (from K322127) + supD-tRNA BEFORE purification</p><p>6) K238013 + B0015 AFTER purification</p><p>7) K238013 + B0015 BEFORE purification</p><p>8) K228000 + RBS AFTER purification</p><p>9) K228000 + RBS BEFORE purification</p><p>10) PCR-product (from K322127) + supD-tRNA BEFORE purification [ligated with T4 ligase]</p><p>11) 2-log | <p>Analytical agarosegel [130711-ligation-control.tif] (1%, 1xTBE, 20 wells, 120 V, 400 mA, 90') for control of ligations from 12-07-11:</p><p>10 µl ligation sample + 2 µl loading dye (6x) -> 12 µl, 10 µl loaded on gel</p><p>5 µl 2-log DNA ladder</p><p>lanes:</p><p>1) 2-log</p><p>2) lacZ + T7-promotor AFTER purification</p><p>3) lacZ + T7-promotor BEFORE purification</p><p>4) PCR-product (from K322127) + supD-tRNA AFTER purification</p><p>5) PCR-product (from K322127) + supD-tRNA BEFORE purification</p><p>6) K238013 + B0015 AFTER purification</p><p>7) K238013 + B0015 BEFORE purification</p><p>8) K228000 + RBS AFTER purification</p><p>9) K228000 + RBS BEFORE purification</p><p>10) PCR-product (from K322127) + supD-tRNA BEFORE purification [ligated with T4 ligase]</p><p>11) 2-log | ||
| - | </p><p><a href="/ | + | </p><p><a href="https://static.igem.org/mediawiki/2011/4/41/130711-ligation-control.jpg" class="image" rel="lightbox"><img alt="130711-ligation-control" src="https://static.igem.org/mediawiki/2011/4/41/130711-ligation-control.jpg" width="400" height="230" /></a></p> |
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d07142011">14-07-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| Line 520: | Line 521: | ||
</p><p>4.) 3b -> PCR-product (from K322127) + supD-tRNA AFTER purification</p><p>5.) 4a -> PCR-product (from K322127) + supD-tRNA BEFORE purification</p><p>6.) 4b -> PCR-product (from K322127) + supD-tRNA BEFORE purification</p><p>7.) 4c -> PCR-product (from K322127) + supD-tRNA BEFORE purification</p><p>8.) 5a -> K238013 + B0015 AFTER purification</p><p>9.) 5c -> K238013 + B0015 AFTER purification | </p><p>4.) 3b -> PCR-product (from K322127) + supD-tRNA AFTER purification</p><p>5.) 4a -> PCR-product (from K322127) + supD-tRNA BEFORE purification</p><p>6.) 4b -> PCR-product (from K322127) + supD-tRNA BEFORE purification</p><p>7.) 4c -> PCR-product (from K322127) + supD-tRNA BEFORE purification</p><p>8.) 5a -> K238013 + B0015 AFTER purification</p><p>9.) 5c -> K238013 + B0015 AFTER purification | ||
</p><p>10.) 6a -> K238013 + B0015 BEFORE purification</p><p>11.) 6b -> K238013 + B0015 BEFORE purification</p><p>12.) 6c -> K238013 + B0015 BEFORE purification</p><p>13.) 7b -> K228000 + RBS AFTER purification</p><p>14.) 7a -> K228000 + RBS AFTER purification</p><p>15.) 7c -> K228000 + RBS AFTER purification | </p><p>10.) 6a -> K238013 + B0015 BEFORE purification</p><p>11.) 6b -> K238013 + B0015 BEFORE purification</p><p>12.) 6c -> K238013 + B0015 BEFORE purification</p><p>13.) 7b -> K228000 + RBS AFTER purification</p><p>14.) 7a -> K228000 + RBS AFTER purification</p><p>15.) 7c -> K228000 + RBS AFTER purification | ||
| - | </p><p>16.) 8a -> K228000 + RBS BEFORE purification</p><p>17.) 8b -> K228000 + RBS BEFORE purification</p><p>19.) 8c -> K228000 + RBS BEFORE purification</p><p>19.) 9a -> PCR-product (from K322127) + supD-tRNA BEFORE purification [ligated with T4 ligase]</p><p><a href="/ | + | </p><p>16.) 8a -> K228000 + RBS BEFORE purification</p><p>17.) 8b -> K228000 + RBS BEFORE purification</p><p>19.) 8c -> K228000 + RBS BEFORE purification</p><p>19.) 9a -> PCR-product (from K322127) + supD-tRNA BEFORE purification [ligated with T4 ligase]</p><p><a href="https://static.igem.org/mediawiki/2011/3/3b/14711_digest-after-miniprep.jpg" class="image" rel="lightbox"><img alt="14711 digest-after-miniprep" src="https://static.igem.org/mediawiki/2011/3/3b/14711_digest-after-miniprep.jpg" width="500" height="178" /></a></p><p><br /></p> |
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d07152011">15-07-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| Line 533: | Line 534: | ||
<p>Colony PCR for the ligation of K322127 and supD-tRNA. Therefore 15 colonies of the plates 3,4 and 9 (from 13-7-11) were resuspended in 50 µl nuclease free water. The experimet failed</p><p>Analytical agarosegel (1%, 1xTBE, 20 wells, 120 V, 400 mA, 90') for control of lthe colony-PCR from 15-7-11: | <p>Colony PCR for the ligation of K322127 and supD-tRNA. Therefore 15 colonies of the plates 3,4 and 9 (from 13-7-11) were resuspended in 50 µl nuclease free water. The experimet failed</p><p>Analytical agarosegel (1%, 1xTBE, 20 wells, 120 V, 400 mA, 90') for control of lthe colony-PCR from 15-7-11: | ||
10 µl ligation sample + 2 µl loading dye (6x) -> 12 µl, 10 µl loaded on gel | 10 µl ligation sample + 2 µl loading dye (6x) -> 12 µl, 10 µl loaded on gel | ||
| - | 5 µl 2-log DNA ladder</p><p>plate 3: </p><p>lanes:</p><p>1. 2-log</p><p>2. positive control</p><p>3-17. colonies from plate 3</p><p>18. positive control</p><p><a href="/ | + | 5 µl 2-log DNA ladder</p><p>plate 3: </p><p>lanes:</p><p>1. 2-log</p><p>2. positive control</p><p>3-17. colonies from plate 3</p><p>18. positive control</p><p><a href="https://static.igem.org/mediawiki/2011/6/64/150711-Gel1-PCR-Platte3.jpg" class="image" rel="lightbox"><img alt="150711-Gel1-PCR-Platte3" src="https://static.igem.org/mediawiki/2011/6/64/150711-Gel1-PCR-Platte3.jpg" width="400" height="307" /></a></p><p>plate 4:</p><p>lanes: </p><p>1. 2-log</p><p>2. negative control</p><p>3-17. samples from the colonies of plate 4</p><p><a href="https://static.igem.org/mediawiki/2011/d/d3/15711_colonyPCR-NK-platte4d-r.jpg" class="image" rel="lightbox"><img alt="15711 colonyPCR-NK-platte4d-r" src="https://static.igem.org/mediawiki/2011/d/d3/15711_colonyPCR-NK-platte4d-r.jpg" width="400" height="429" /></a> |
</p><p><br /> | </p><p><br /> | ||
| - | plate 9: </p><p>lanes:</p><p>1. 2-log</p><p>2-17. samples from colonies of plate 9</p><p><a href="/ | + | plate 9: </p><p>lanes:</p><p>1. 2-log</p><p>2-17. samples from colonies of plate 9</p><p><a href="https://static.igem.org/mediawiki/2011/9/94/15711-colonyPCR_platte9d-r.jpg" class="image" rel="lightbox"><img alt="15711-colonyPCR platte9d-r" src="https://static.igem.org/mediawiki/2011/9/94/15711-colonyPCR_platte9d-r.jpg" width="400" height="336" /></a></p><p><br /></p> |
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d07192011">19-07-2011</span></h2> |
<h3> <span class="mw-headline">Results</span></h3> | <h3> <span class="mw-headline">Results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| Line 548: | Line 549: | ||
<p>Restriction digest of K238013+B0015 with S, P and K228000 (T7-Polymerase) + RBS with X, P</p><p>Restriction digest (total volume: 30 µl; DNA: 1 µg; Enzyme 1 µl each; 3 µl NEB 4 buffer (10x); incubation: 1 h @ 37°C)</p><p>The used DNA-templates were 8a (14-7-11) and 6c (14-7-11) | <p>Restriction digest of K238013+B0015 with S, P and K228000 (T7-Polymerase) + RBS with X, P</p><p>Restriction digest (total volume: 30 µl; DNA: 1 µg; Enzyme 1 µl each; 3 µl NEB 4 buffer (10x); incubation: 1 h @ 37°C)</p><p>The used DNA-templates were 8a (14-7-11) and 6c (14-7-11) | ||
</p><p><br /> | </p><p><br /> | ||
| - | preparative agarosegel (1%, 1xTAE, 8 wells, 120 V, 400 mA, 90'):</p><p>30 µl restriction digest + 6 µl loading dye (6x) -> 35 µl loaded on gel</p><p>lanes:</p><p>1.) 2 log- DNA ladder 7 µl</p><p>2.) K22800 + RBS</p><p>3.) K238013 + B0015</p><p>4.) K322127 from an earlier restriction digest</p><p>-> DNA extraction from gel via mi-Gel-extraction kit from metabiom</p><p><a href="/ | + | preparative agarosegel (1%, 1xTAE, 8 wells, 120 V, 400 mA, 90'):</p><p>30 µl restriction digest + 6 µl loading dye (6x) -> 35 µl loaded on gel</p><p>lanes:</p><p>1.) 2 log- DNA ladder 7 µl</p><p>2.) K22800 + RBS</p><p>3.) K238013 + B0015</p><p>4.) K322127 from an earlier restriction digest</p><p>-> DNA extraction from gel via mi-Gel-extraction kit from metabiom</p><p><a href="https://static.igem.org/mediawiki/2011/6/60/19711_verdau_t7pol-RBS_K238013-B0015_K322127.jpg" class="image" rel="lightbox"><img alt="19711 verdau t7pol-RBS K238013-B0015 K322127" src="https://static.igem.org/mediawiki/2011/6/60/19711_verdau_t7pol-RBS_K238013-B0015_K322127.jpg" width="250" height="343" /></a> <a href="/wiki/images/5/52/19711_verdau_t7pol-RBS_K238013-B0015_K322127_nachher.jpg" class="image" rel="lightbox"><img alt="19711 verdau t7pol-RBS K238013-B0015 K322127 nachher" src="/wiki/images/5/52/19711_verdau_t7pol-RBS_K238013-B0015_K322127_nachher.jpg" width="250" height="338" /></a></p><p><br /></p> |
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d07202011">20-07-2011</span></h2> |
<h3> <span class="mw-headline">results</span></h3> | <h3> <span class="mw-headline">results</span></h3> | ||
<div class="results"> | <div class="results"> | ||
| Line 562: | Line 563: | ||
<h4> <span class="mw-headline" id="Transformation_9">Transformation</span></h4> | <h4> <span class="mw-headline" id="Transformation_9">Transformation</span></h4> | ||
<p>transformation (40 µl competent cells; 1 µl DNA; Electroporation at 1510 V; incubation in Soc-Medium for 1 h @ 37°C, 50 µl on Amp-agar plate)</p><p>analytical agarosegel (1%, 1xTBE, 8 wells, 120 V, 400 mA, 90') | <p>transformation (40 µl competent cells; 1 µl DNA; Electroporation at 1510 V; incubation in Soc-Medium for 1 h @ 37°C, 50 µl on Amp-agar plate)</p><p>analytical agarosegel (1%, 1xTBE, 8 wells, 120 V, 400 mA, 90') | ||
| - | </p><p>10 µl DNA + 2 µl loading dye -> 10 µl on gel</p><p>Lanes:</p><p>1.) 7 µl 2-log DNA ladder</p><p>2.) K228000-RBS + K238013 - B0015 (upper band in preparative gel)</p><p>3.) K228000-RBS + K238013 - B0015 (lower band in preparative gel)</p><p>4.) K322127-tRNA</p><p><a href="/ | + | </p><p>10 µl DNA + 2 µl loading dye -> 10 µl on gel</p><p>Lanes:</p><p>1.) 7 µl 2-log DNA ladder</p><p>2.) K228000-RBS + K238013 - B0015 (upper band in preparative gel)</p><p>3.) K228000-RBS + K238013 - B0015 (lower band in preparative gel)</p><p>4.) K322127-tRNA</p><p><a href="https://static.igem.org/mediawiki/2011/1/1b/20711_ligation_analytisch.jpg" class="image" rel="lightbox"><img alt="20711 ligation analytisch" src="https://static.igem.org/mediawiki/2011/1/1b/20711_ligation_analytisch.jpg" width="250" height="301" /></a></p><p><br /></p> |
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d07212011">21-07-2011</span></h2> |
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
<div class="cloning"> | <div class="cloning"> | ||
<h4> <span class="mw-headline" id="Restriction_4">Restriction</span></h4> | <h4> <span class="mw-headline" id="Restriction_4">Restriction</span></h4> | ||
<p>control restriction digest with the Enzymes E, P of 2b (=ligation product lacZ+T7 promotor), 6c (=ligation product K238013+B0015) and 8a (ligation product K228000+RBS). </p><p>Restriction digest (total volume: 30 µl; DNA: 0.5 µg; Enzyme 1 µl each (E, P); 3 µl NEB 4 buffer (10x); incubation: 1 h @ 37°C)</p><p>analytical gelelektrophoresis (1%, 1x TBE, 12 wells, 120 V, 400 mA, 90´)</p><p>lanes:</p><p>1.) 2b ( =ligation product lacZ+T7-Promotor) | <p>control restriction digest with the Enzymes E, P of 2b (=ligation product lacZ+T7 promotor), 6c (=ligation product K238013+B0015) and 8a (ligation product K228000+RBS). </p><p>Restriction digest (total volume: 30 µl; DNA: 0.5 µg; Enzyme 1 µl each (E, P); 3 µl NEB 4 buffer (10x); incubation: 1 h @ 37°C)</p><p>analytical gelelektrophoresis (1%, 1x TBE, 12 wells, 120 V, 400 mA, 90´)</p><p>lanes:</p><p>1.) 2b ( =ligation product lacZ+T7-Promotor) | ||
| - | </p><p>3.) 6c (=ligation product K238013 B B0015)</p><p>5.) 8A (=ligation product K228000 + RBS)</p><p>6.) 2-log DNA ladder</p><p><a href="/ | + | </p><p>3.) 6c (=ligation product K238013 B B0015)</p><p>5.) 8A (=ligation product K228000 + RBS)</p><p>6.) 2-log DNA ladder</p><p><a href="https://static.igem.org/mediawiki/2011/a/a7/21711_kontrollverdau_2b_6c_8a.jpg" class="image" rel="lightbox"><img alt="21711 kontrollverdau 2b 6c 8a" src="https://static.igem.org/mediawiki/2011/a/a7/21711_kontrollverdau_2b_6c_8a.jpg" width="250" height="301" /></a></p> |
</div> | </div> | ||
<h3> <span class="mw-headline">Other Work</span></h3> | <h3> <span class="mw-headline">Other Work</span></h3> | ||
| Line 577: | Line 578: | ||
</p><p>- 5ml overnight culture of 218 (pBADlacZ) with Ampicillin</p><p>- 5 ml overnight culture of BM28 with Kanamycin </p><p>-> both cultures are used for gelrite experiments</p> | </p><p>- 5ml overnight culture of 218 (pBADlacZ) with Ampicillin</p><p>- 5 ml overnight culture of BM28 with Kanamycin </p><p>-> both cultures are used for gelrite experiments</p> | ||
</div> | </div> | ||
| - | + | <br> | |
| - | <h2><span class="mw-headline" id=" | + | <h2><span class="mw-headline" id="d07222011">22-07-2011</span></h2> |
<h3> <span class="mw-headline">Cloning</span></h3> | <h3> <span class="mw-headline">Cloning</span></h3> | ||
<div class="cloning"> | <div class="cloning"> | ||
| Line 585: | Line 586: | ||
<h4> <span class="mw-headline" id="Restriction_5">Restriction</span></h4> | <h4> <span class="mw-headline" id="Restriction_5">Restriction</span></h4> | ||
<p>control regristiction digest of 10 a-c (ligation product K322127 + tRNA) and 11 a-c (ligation product K228000-RBS + K238013+B0015) with E, P</p><p>Restriction digest (total volume: 30 µl; DNA: 0.3 µg; Enzyme 1 µl each (E, P); 3 µl NEB 4 buffer (10x); incubation: 1 h 10` @ 37°C, inactivation 20´@ 80°C)</p><p>analytical gelelectrophoresis (1%, TAE 1x, 12 wells, 120 V, 400 mA, 120`)</p><p>lanes:</p><p>1.) 2-log ( 1 µl 2-log, 1 µl loading buffer (6x), 10 µl water) -> 10 µl loaded on gel</p><p>2.) 10 a</p><p>3.) 10 b</p><p>4.) 10 c</p><p>5.) 11 a | <p>control regristiction digest of 10 a-c (ligation product K322127 + tRNA) and 11 a-c (ligation product K228000-RBS + K238013+B0015) with E, P</p><p>Restriction digest (total volume: 30 µl; DNA: 0.3 µg; Enzyme 1 µl each (E, P); 3 µl NEB 4 buffer (10x); incubation: 1 h 10` @ 37°C, inactivation 20´@ 80°C)</p><p>analytical gelelectrophoresis (1%, TAE 1x, 12 wells, 120 V, 400 mA, 120`)</p><p>lanes:</p><p>1.) 2-log ( 1 µl 2-log, 1 µl loading buffer (6x), 10 µl water) -> 10 µl loaded on gel</p><p>2.) 10 a</p><p>3.) 10 b</p><p>4.) 10 c</p><p>5.) 11 a | ||
| - | </p><p>6.) 11 b</p><p>7.) 11 c</p><p><a href="/ | + | </p><p>6.) 11 b</p><p>7.) 11 c</p><p><a href="https://static.igem.org/mediawiki/2011/6/6b/110722_Gel_Right_1.jpg" class="image" rel="lightbox"><img alt="110722 Gel Right 1" src="https://static.igem.org/mediawiki/2011/6/6b/110722_Gel_Right_1.jpg" width="250" height="225" /></a></p> |
</div> | </div> | ||
</div> | </div> | ||
Latest revision as of 02:39, 22 September 2011
Cloning Part I
People: Wolfgang, Tobi, Nico, Katharina, Flo
11-05-2011
Other Work
We request the following parts from the registry:
| Part | Team | Library | Plate | Well | Plasmid | Resistance |
|---|---|---|---|---|---|---|
| BBa_K238013 | iGEM09_KULeuven | 2009 Submissions | Shipment: 00413 | 1 | pSB1A2 | A |
| BBa_K228000 | iGEM09_PKU_Beijing | 2009 Submissions | Shipment: 00399 | 1A | pSB1A2 | A |
| BB a_K322127 | iGEM10_Edinburgh | Submissions | Shipment: 00694 | 6 | pSB1C3 | C |
| BBa_K228823 | iGEM09_PKU_Beijing | 2009 Submissions | Shipment: 00401 | 11B | pSB4K5 | K |
| Bba_K322119 | iGEM10_Edinburgh | unknown | unknown | unknown | unknown | |
| Bba_K322115 | iGEM10_Edinburgh | unknown | unknown | unknown | unknown |
12-05-2011
Autoclave broken
We are not able to prepare any media or sterile materials. Should be working next week again (Andrea is going to tell us).
-> No agar plate/medium preparation possible right now.
But all other necessary stuff seems to be available (for electroporation, competent cells, miniprep-kit, DH5alpha strain, BL21 strain).
19-05-2011
Autoclave working
Other Work
We start preparing Agar-LB Plates with Amp and Kan, as well as normal LB medium.
20-05-2011
Cloning
Transformation
Transformation of all red-light sensor parts separately. DH5 alpha cells were transformed using electroporation (V = 1600V) with following constructs:
Part alias : Part Name - Plasmid - Year of Distribution, Plate Number
iGEM R1 : R0082 pSB1A3 2010, P1
iGEM R2 : I732017 pSB1A2 2010, P2
iGEM R3 : I15010 pSB2K3 2010, P3
iGEM R4 : K098010 pSB4C5 2010, P3
Each electroporation was performed with 1.5 µl BioBrick plasmid (each disolved in 10 µl ddH2O before) and 40 µl competent DH5 alpha cells according to the protocol kindly provided by Andrea Mueckl.
Afterwards cells were incubated in 1 ml SOC medium for 1.5 h at 37 °C, 200 rpm and subsequently plated on LB-agar plates containing antibiotics (50 µl, 100 µl, 200 µl). They were incubated at 37 °C over night and then stored at 4 °C.
23-05-2011
Results
Transformation of all red sensor parts failed. This might have been due to a wrong electroporation protocol.
Cloning
Transformation
Transformation of one red part sensor (iGEM R0082) with slightly modified protocol: electroporation was performed at 1500 V with 1 µl Plasmid (part was previously disolved in 10 µl ddH2O)and 40 µl competent DH5 alpha cells. Otherwise same procedure as before.
Other Work
Making electrocompetent cells according to Andrea's Mueckl protocol.
24-05-2011
Results
Transformation of R0082 resulted in 3 colonies.
Other Work
Inoculation of over-night culture (20 ml) of DH5 alpha cells in Luria-Media for electrocompetent cells.
Inoculation of over-night culture of one clone picked from the successful second transformation.
We asked Dr. J. Winter for heatresistant E. coli. We should get them tomorrow after lunch on a plate and can then culture them.
25-05-2011
Results
The clone picked yesterday turned out as the right one.
Dr. J. Winter wants us to come back tomorrow, since bacteria seem to be contaminated
Cloning
MiniPrep
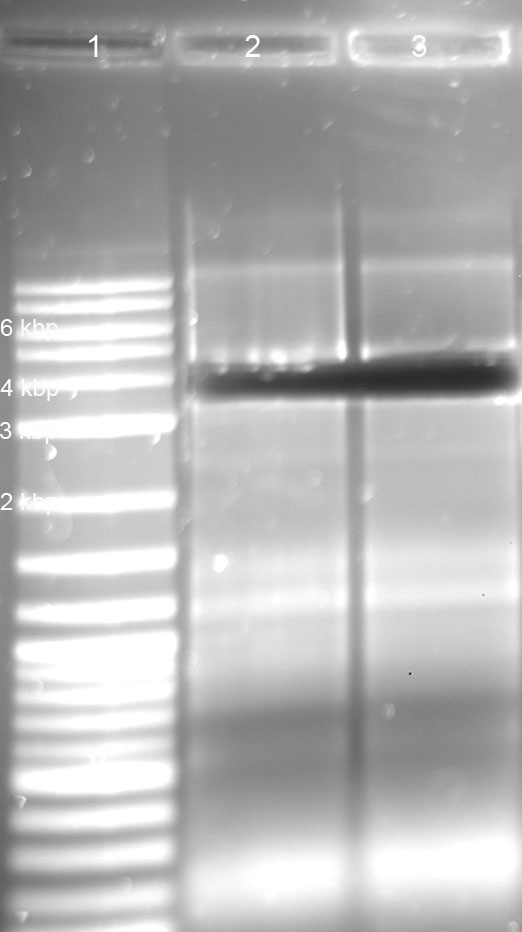
Testing of the clone: Mini-Prep using Zymoresearch DNA Kit, with subsequent digestion with EcoRI and PstI in NEB-Buffer 4. Afterwards loading ontol Agarose gel (expected fragments 0.1 kbp (= R0082) and 2,0 kbp (= pSB1A2))
Other Work
Continue making cells electrocompetent.
26-05-2011
Results
Again: Dr. J. Winter wants us to come back next day, since bacteria seem to be contaminated.
Cloning
Transformation
Transformation of
Part alias : Part Name - Plasmid - Year of Distribution, Plate Number
iGEM R2 : I732017 pSB1A2 2010, P2
iGEM R3 : I15010 pSB2K3 2010, P3
iGEM R4 : K098010 pSB4C5 2010, P3
We were using 1510 V and 1 µl DNA for transformation. The DNA was just added to the cells and gently mixed by stirring it with the pipette tip. The protocol used was the one from Andrea Meyer. Of the 1 ml SOC culture, 50 µl were spread out directly on Agar plates. Rest was spun down (2000 rpm, 1 min) and pellet was resuspended in 100 to 150 µl. Subsequently, 50 µl were spread out.
27-05-2011
Results
There are successful clones from transformed cells with iGEM I732017 and iGEM K098010, but no clones from iGEM I15010 at all
Again: Dr. J. Winter wants us to come back next week, as there is still contamination in the culture.
Cloning
Transformation
Transformation of:
Part alias : Part Name - Plasmid - Year of Distribution, Plate Number
iGEM R3 : I15010 pSB2K3 2011, P3
iGEM tRNA : K228001, pSB1A2 2011, P4
iGEM Term : B0015 pSB1AK3 2011, P1
iGEM RBS : J44001 pSB1A2 2011, P1
iGEM T7 : I712074 pSB1AK8 2011, P1
Since the transformation with iGEM I15010 did not work, we increased the amount of plasmid to 2 µl. Afterwards the procedure was performed as mentioned above and the cells were incubated over night (15 h) at 37 °C and cooled until monday at 4 °C.
30-05-2011
Results
We have successful clones from transformed cells with iGEM K228001 (tRNA), iGEM B0015 (Term), iGEM J44001 (RBS), iGEM I712074 (T7).
Finally, we received BM28 cells from Dr. J. Winter (there is an existing paper in JBC from her about the bacteria).
Cloning
Transformation
Transformation of I15010 once more using BL21 and inducing pSB2K3 plasmid using 1 mM IPTG in SOC-Medium.
Other Work
Inoculation of over-night cultures of clones from iGEM K228001 (tRNA), iGEM B0015 (Term), iGEM J44001 (RBS), iGEM I712074 (T7), iGEM I732017 und iGEM K098010
31-05-2011
Results
The transformation of iGEM I15010 (R3) did not work.
We measured plasmid concentrations from MiniPreps (see below) using nano-drop:
- iGEM I712074 (T7): 62 ng/µl
- iGEM K098010 (R4): 82,5 ng/µl
- iGEM K228001 (tRNA): 98 ng/µl
- iGEM I732017 (R2): 94 ng/µl
- iGEM B0015 (Term): 141 ng/µl
- iGEM J44001 (RBS): 108 ng/µl
- iGEM R0082 (R1): 59 ng/µl
Cloning
MiniPrep
Plasmid Isolation (MiniPrep) from all over night cultures was conducted and the concentrations were measured.
01-06-2011
Results
A SDS-PAGE was conducted from the cultures induced with IPTG on 50-05-2011. The cells were boiled for 5 min at 95 °C in loading buffer. The gel ran inconclusive, next time there should be alonger run with higher voltage. Only 1/3 of the lane was used by the peptides. It had a very bad resolution. However, the dying step worked properly.
Cloning
Digestion
- A restriction digestion of the parts gained by MiniPrep was conducted using EcoRI and PstI in NEB-Buffer 4. Afterwards, they were applied onto an agarose gel.
- Properties of the gel run with the digested samples: 100 V, 1 h 10 Min, 1 % agarose.
- SYBR Gold staining: 2 µl on shaking plate for 30 mins at room temperature (RT).
Transformation
Transformation of DH5 alpha with:
iGEM I15010 (R3) (see above) on ampicilin (A, Amp), chloramphenicol (C, Cm) and kanamycin (K, Kan) containing resistance plates;
iGEM High Copy Plasmid, Plate 1 2011 1G, psB1A3 with insert: BBa_J04450, Resistance: A;
iGEM Low Copy Plasmid, Plate 1 2011 3C, psB3C5 with insert: BBa_J04450, Resistance: C.
They were plated on agar plates carrying the appropriate antibiotic resistances and incubated at 37 °C over night.
03-06-2011
Results
Another SDS-PAGE gel run again inconclusive. Furthermore during the run, there was a loss of electrical power because of some blackout in the building.
Transformed cells grew very well over night, with a few clones of iGEM I15010 (R3) on kanamycin resistance plate found. This is evidence for a contamination, because the incubator did not cool down to 4 °C over thursday.
The transformed plasmid-cells grew also very good - now featured in red.
Cloning
Digestion
An agarose gel was run with digestions from yesterday, using the following settings: 90 V, 1 h 30 Min, 1 % agarose.
SYBR Gold staining: 2 µl on shaking plate for 30 mins at RT.
06-06-2011
Results
The agarose gel was again inconclusive. There may be too less material, since no bands around 100 bp could be detected at all.
Cloning
Digestion
Another digestion was conducted and analyzed on an agaorse gel. It ran with newly digested and undigested plasmid as control: 100 V, 1 h 30 Min, 1 % agarose.
Transformation
Transformation of I15010 (R3) in E. coli D1210
Other Work
Over-night culture of the following parts were inoculated:
iGEM High Copy Plasmid, Plate 1 2011 1G, psB1A3 with insert: BBa_J04450, Resistance: A;
iGEM Low Copy Plasmid, Plate 1 2011 3C, psB3C5 with insert: BBa_J04450, Resistance: C.
07-06-2011
Results
The transformation of I15010 (R3) finally worked using E. coli D1210 cells.
MiniPrep of the over night culture of iGEM Low Copy Plasmid, Plate 1 2011 3C, psB3C5 with insert: BBa_J04450, Resistance: C resulted in 101 ng/µl in a total volume of 100 µl.
We have run out of DNA-Plasmid Preparation Kit. No more preparations are possible during the next days, until we have a new kit!
A new digestion was conducted. The subsequent gel still was inconclusive.
Cloning
Digestion
The digestion was repeated. In order to have a more sensitive detection of DNA on the gel, we now use ethidium bromide for staining of the gel. The following settings are used: 2 % Agarose with ethidium bromide, 90 V, 1.5 h.
Digestion of 2 µg Plasmid DNA with high fidelity 1.: EcoRI and PstI enzymes and 2.: just with PstI was conducted.
08-06-2011
Results
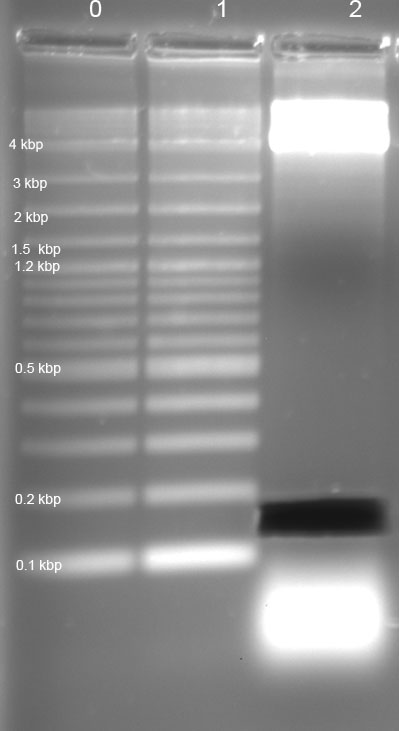
gel: R1, R2, R4 tRNA und Term seem ok; T7 promotor (46bp) and RBS (15bp) could not be detected ---> PAGE necessary!
Cloning
Digestion
1.5 % Agarose with Sybr Gold (Dilution: 1 µl/10ml), 90V, 1.0 h
09-06-2011
Results
gel: RBS, T7 digestion -> PAGE not successful;
DNA Conc. after MiniPrep:
BBa_K238013 on pSB1A2: 49ng/ul
BBa_K228000 on pSB1A2: 204 ng/ul
BBa_K322127 on pSB1C3: 168 ng/ul
Cloning
MiniPrep
MiniPrep of BBa_K238013 BBa_K228000 BBa_K322127
Other Work
Over Night Culture of R3 clone
Testing
SDS Page
10% PAGE of RBS and T7
10-06-2011
Results
gel:
DNA Conc. after MiniPrep:
R3 188 ng/ul
Cloning
MiniPrep
MiniPrep of R3
Comparison of "new" (ordered) enzymes vs. "old" ones
14-06-2011
Results
Other Work
Comparison of "new" (ordered) enzymes vs. "old" ones - running the gel (1% agarose, 100-150ml Gel) (Cut R4, R2)
16-06-2011
Cloning
Cloning of the red light sensor with parts R1,R2,R4 (see cloning draft)
ONC of part containing colonies in 5ml liquid culture
17-06-2011
Results
NO COLONIES of first ligation ->problem: wrong enzymes used! (forgot to cut the linear fragment in the second step with pstI)
gel: inconclusive
Cloning
Ligation
Agarose gel of ligation products to evaluate ligation problems
MiniPrep
MiniPreps of Parts
20-06-2011
Results
Gel of samples collected during the red light sensor assembly: FILE
Cloning
-Cloning of the red light sensor with parts R1,R2,R4 (see cloning draft)
Other Work
-Backup Plates of existing clones
21-06-2011
Results
-Some strange looking colonies of red light sensor transformation
Cloning
-ONC of part containing colonies and red light sensor construct colonies in 5ml liquid culture
Cloning
-Cloning of the blue sensor (only step1) with parts K228000, RBS (see cloning draft)
22-06-2011
Results
-nice colonies of red light sensor transformation
Cloning
-Cloning of the blue sensor (step2) with parts: Y44001 (RBS) + K22800 = A1 and B 0015 (Term)
-Cloning of the red light sensor with lacZ (R2)
Digestion
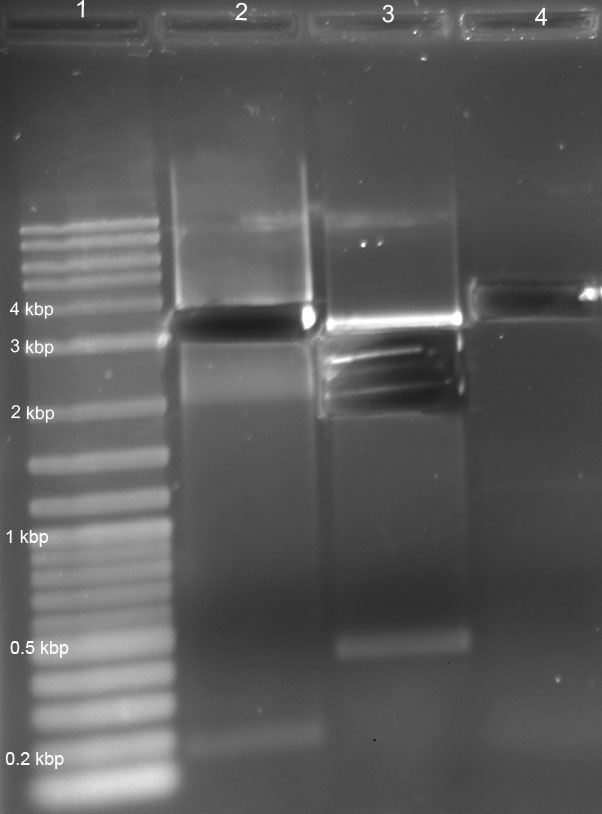
- gel run of all products (110 V, 1%, 1h):
lanes: 2log ladder, RBS undigested (=ud), RBS digested (dig), K22800 ud, K22800 dig, A1 ud, A1 dig, Term ud, Term dig, L1 ud, L1 dig, L2 ud, L2 dig, R2 ud, R2 dig
6 µl each lane
gel: 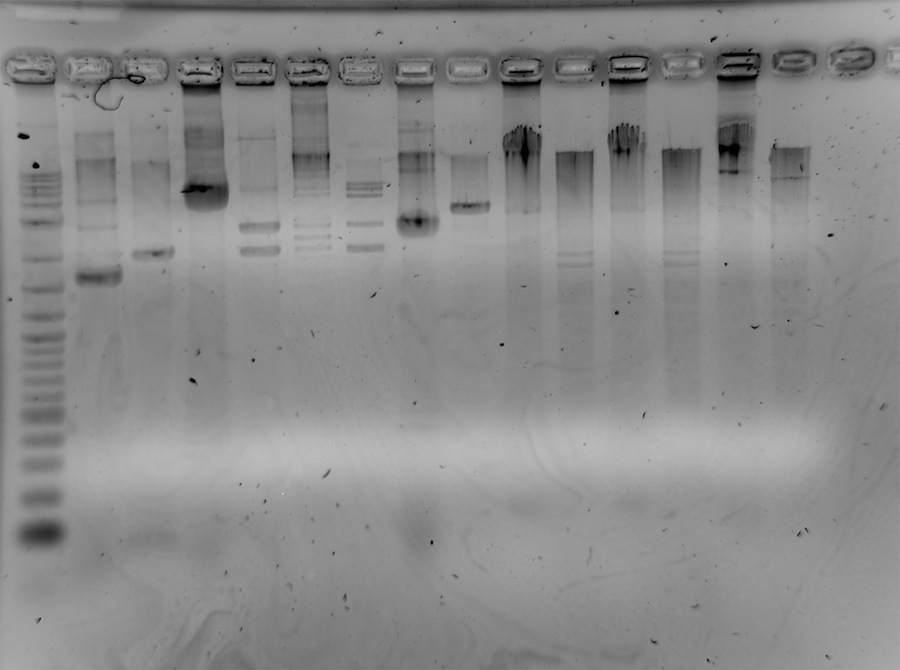
Transformation
-Transformation of both ligations (red and blue light) and incubation on plates over night:
- blue light on amp and kan
- red light I on amp plate not protected from light, lightning during electroporation
- red light II on amp 1 plate protected from light, 1 plate not protected from light
MiniPrep
- plasmid isolation of all red light sensor parts and the successfully transformed colonies from 21.06.11 concentrations in ng/µl
R1 93.5 R2 108.0 R3 37.0 R4 263.0 RBS 47.5 tRNA 154.0 Term 65.0 Ligation of transformed colony (21.6.11) I 95.9 Ligation of transformed colony (21.6.11) II 63.5 T7 106.0 high copy plasmid 142.0 low copy plasmid 66.5
24-06-2011
Other Work
New agarose plates
27-06-2011
Other Work
Design of new cloning strategy together with instructors
28-06-2011
Cloning
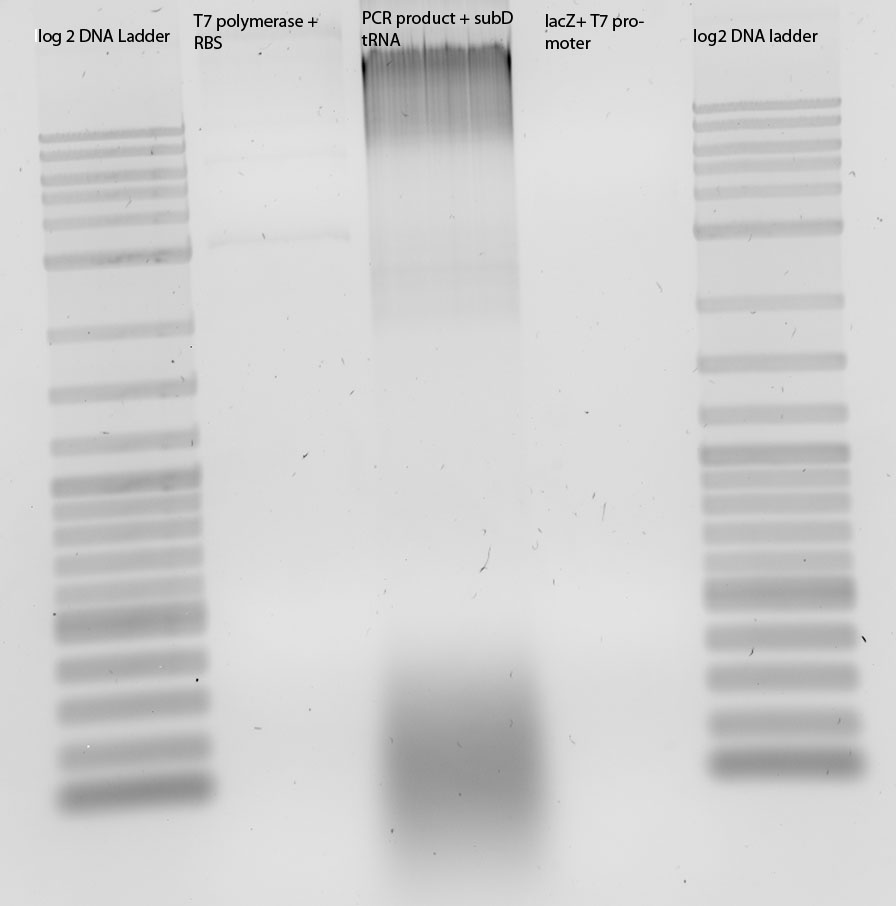
Digestion (EcoRIxPstI) and ligation of K238013, tRNA in pSB3C5, blue light construct (j44001-K22800-B0015), Reporter construct (T7-R2)
29-06-2011
Results
Bad gel
Cloning
Digestion (EcoRIxPstI) of K238013, tRNA in pSB3C5, blue light construct (j44001-K22800-B0015), Reporter construct (T7-R2)
30-06-2011
Results
Agarose Gel of K238013 (Insert: 86bp), tRNA in pSB3C5 (Insert: 136bp), blue light construct (j44001-K22800-B0015) (Insert: 3,2kb), Reporter construct (T7-R2) (Insert: 3,1kb)
In T7-R2 only T7 and in blue light construct only B0015? Maybe there is also the right band. Do our ligations work? (see cloning road maps...)
Cloning
Digestion
Digestion (ExP) of B0015, pSB1A3 (not needed), pSB3C5
Agarose Gel of K238013, tRNA in pSB3C5, blue light construct (j44001-K22800-B0015), Reporter construct (T7-R2)
01-07-2011
Cloning
Amplification
PCR to amplify certain parts out of the red light sensor of edinburgh: K322123, K322124, ompR:
PCR-Program:
1) Initial Denaturation: 96°C, 4'
2) Touchdown (8x)
Denat.: 96°C, 30 s Annealing: 65°C -> 61,5°C; -0,5°C/cycle, 20 s Extension: 72°C, 4'
3) Constant Temp. (22x)
Denat.: 96°C, 30 s Annealing: 61°C, 20 s Extension: 72°C, 4'
4) Final Extension: 72°C, 8'
5) Hold: 4°C, oo
PCR-mix (8x) (Taq-PCR-Kit; NEB):
water (autoclaved): 326 µl
standard buffer (10x): 40 µl
dNTPs (10mM): 8 µl
Taq-Pol: 2 µl
47 µl per reaction + 1 µl of each primer + 1 µl DNA -> 50 µl reaction volume
samples:
1) negative control: primer (10 µM) + water
2) DNA: 1,5 ng; primer (10 µM)
3) DNA: 15 ng; primer (10 µM)
4) DNA: 1,5 ng; primer (5 µM)
5) DNA: 15 ng; primer (5 µM)
6) DNA: 1,5 ng; primer (1 µM)
7) DNA: 15 ng; primer (1 µM)
Other Work
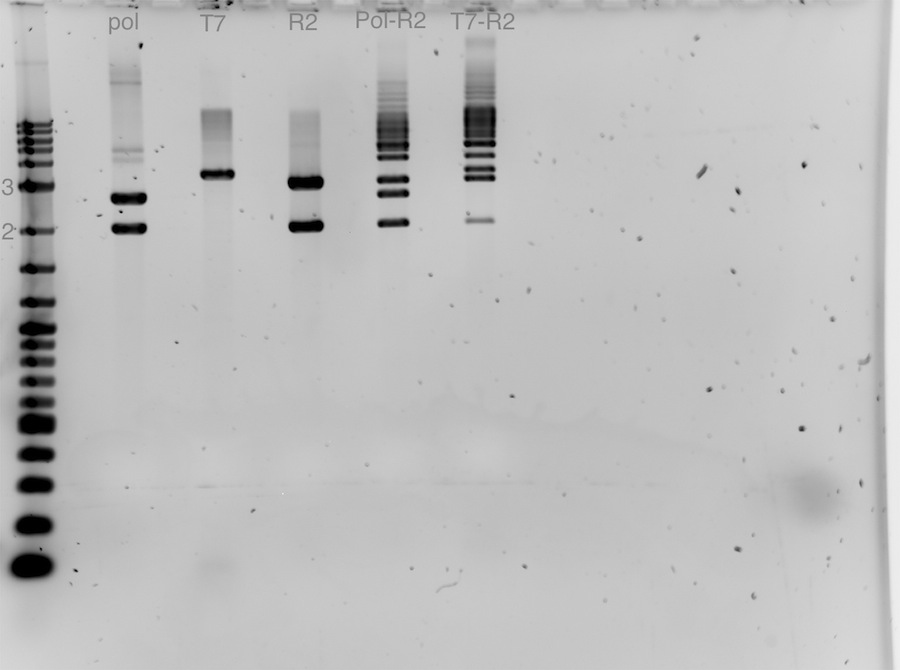
Agarose Gel of ladder, R2, T7, Pol, lac-Pol(R2-Pol), T7-R2, Term. low copy 1:1, Term low copy 3:1, ladder 2 runs with different conditions: 120 V, 400 mA, 1.5 h<-> 80 V, 120 mA, 1.5h
Making of electrocompetent cells of DH5alpha and BM28
04-07-2011
Results
Cloning
Agarose
Agarose Gel of ladder, R2, T7, Pol, lac-Pol(R2-Pol) ligation, T7-R2 ligation
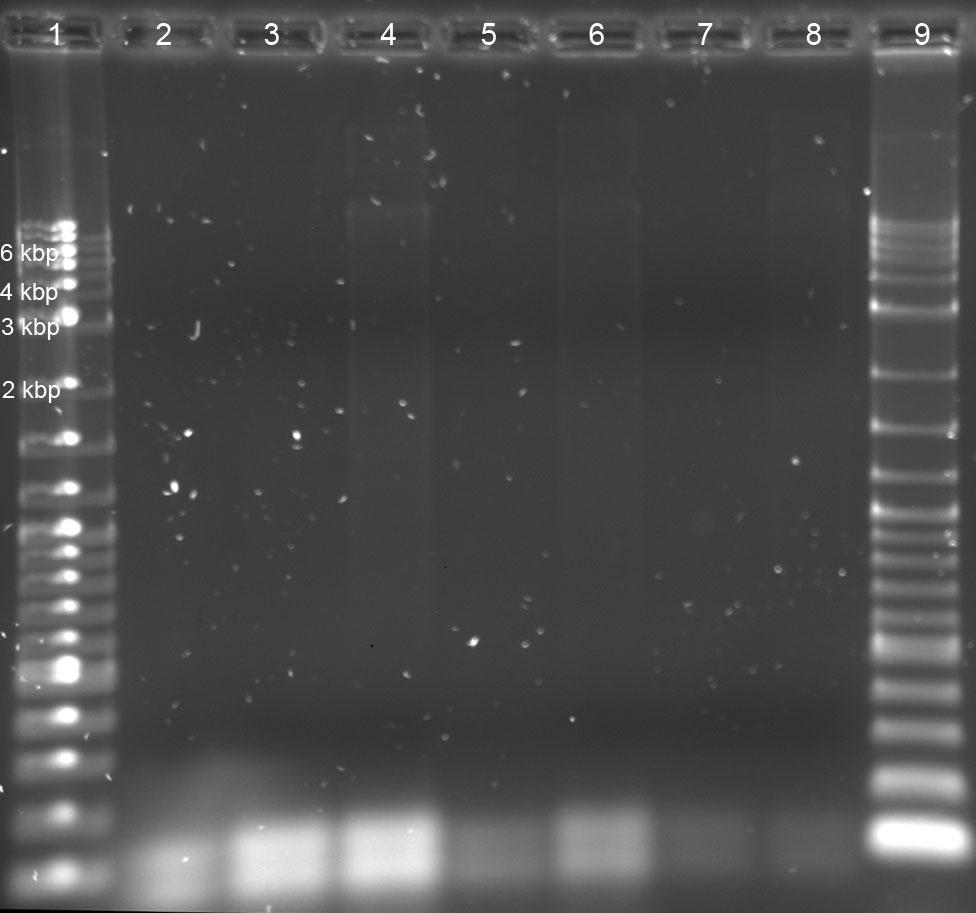
analytical agarose gelelectrophoresis of the PCR samples from 01-07-11: (1%, 1x TBE, 120 V, 400 mA, 90 min)
10 µl PCR sample + 2 µl loading buffer -> 10 µl per well
lanes:
1.) 2-log
2) negative control: primer (10 µM) + water
3) DNA: 1,5 ng; primer (10 µM)
4) DNA: 15 ng; primer (10 µM)
5) DNA: 1,5 ng; primer (5 µM)
6) DNA: 15 ng; primer (5 µM)
7) DNA: 1,5 ng; primer (1 µM)
8) DNA: 15 ng; primer (1 µM)
9) 2-log
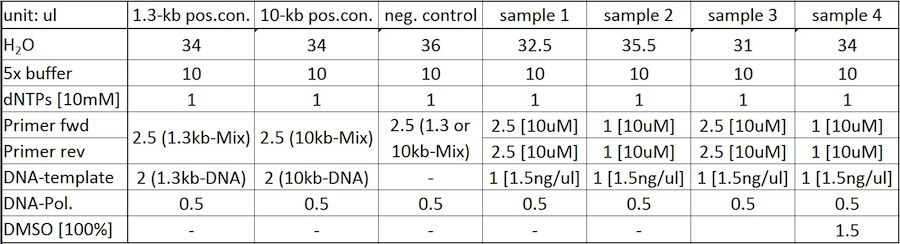
05-07-2011
Cloning
Amplification
repetition of PCR to amplify certain parts out of the red light sensor of edinburgh: K322123, K322124, ompR:
Phusion High-Fidelity PCR-Kit (NEB/Finnzymes)
PCR-Program:
1) Initial Denaturation: 98°C, 4'
2) 30 cycles of:
Denat.: 98°C, 30 s Annealing: 60°C, 15 s Extension: 72°C, 2´ 30 s
3) Final extension: 72°C, 8'
4) Hold: 4°C, oo
Samples
PCR-samples were prepared with HF- and GC-buffer, respectively. End volume of the reaction setups was 50 ul.
DMSO-concentration was 100%; primer concentration was 10 uM, which equals an end conc. of 500 nM (2.5ul) or 200 nM (1ul).
DNA-template [150 ng/ul] was diluted 1:100 to a conc. of 1.5 ng/ul -> 1 ul of dilution was used per reaction.
Samples were prepared as follows (same setup for HF- and GC-buffer):
06-07-2011
Results
HF-buffer samples gel analysis:
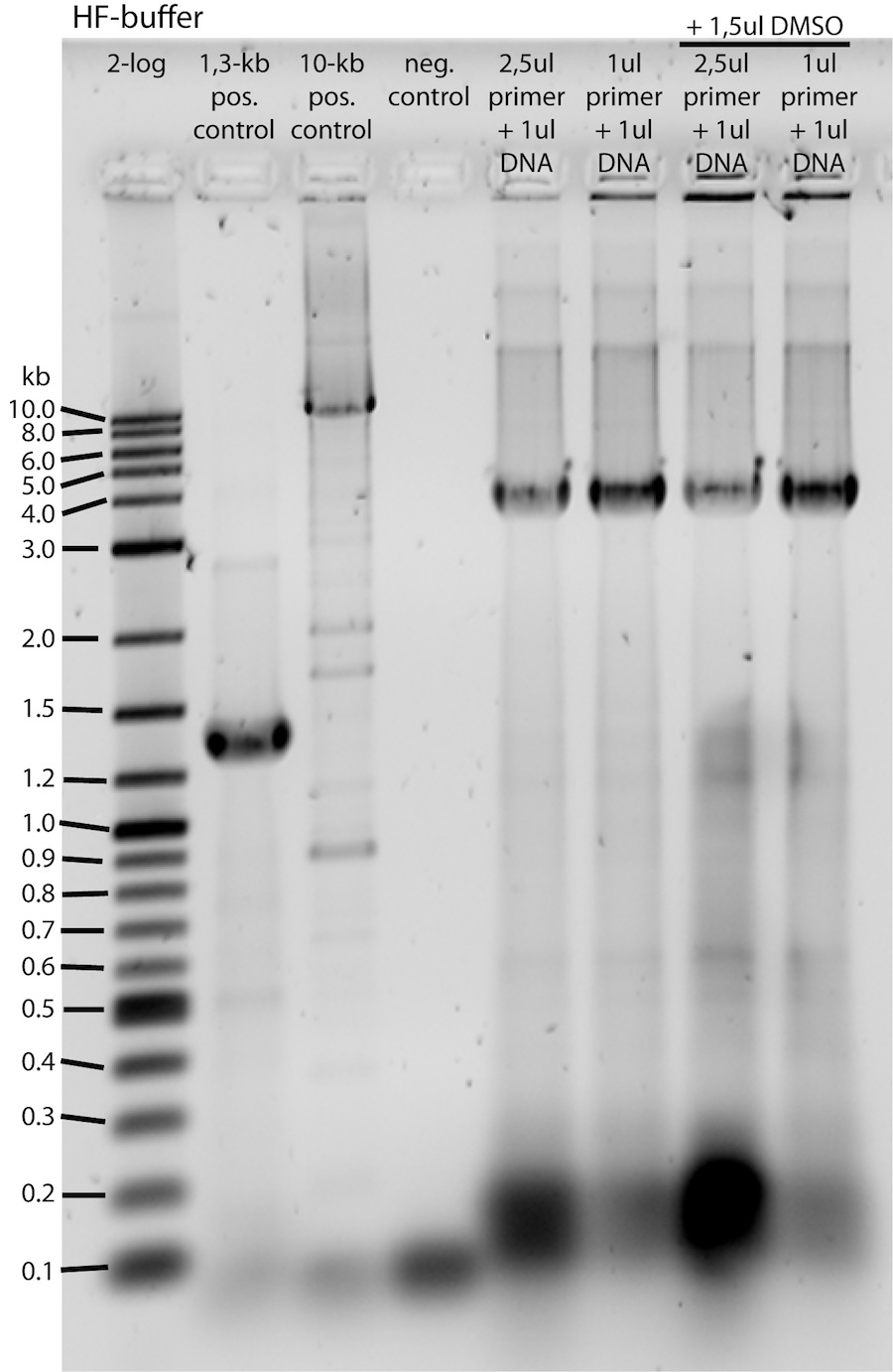 -> PCR seems to have been successful, amplicon is at about 4kb!
-> PCR seems to have been successful, amplicon is at about 4kb!
QC-buffer samples gel analysis (before and after cut-out):
 -> PCR also successful, cut-out of 4 bands, DNA prep and pooling samples
-> PCR also successful, cut-out of 4 bands, DNA prep and pooling samples
Cloning
Digestion
Digestion of J44001, K228000
Preparative gel run with the digested parts (1% agarose, 1xTAE, 120 V 400 mA, 1.5 h)
Isolation of the bands out of the gel
Preparation with a gel isolation kit (freeze ´n squeeze, bio-rad)
work-up of PCR-run (05-07-2011): gel analysis (1% agarose, 1xTBE, 120 V, 400 mA, 1.5 h) of HF-buffer-samples
gel analysis (1% agarose, 1xTBE, 120 V, 400 mA, 1.5 h) of GC-buffer-samples
isolation of right bands out of the GC-gel
DNA preparation with a gel isolation kit (freeze ´n squeeze, bio-rad)
restriction digest with EcoRI and SpeI
MiniPrep
mini-prep of culture (4 ml) from DH5alpha (05-07-2011) (qiagen qiaprep spin miniprep kit) -> 82 ng/µl in 50 µl elution buffer
restriction digest of 0.5 µg DNA with EcoRI and PstI
gel analysis of restriction digest (1% agarose, 1xTAE, 120 V, 400 mA, 1.5 h)
07-07-2011
Cloning
Ligation
Ligation (total volume: 50 µl; 1.5 µg of each DNA sample; 5 µl T4-ligase buffer (10x); 1 µl Quick-Ligase; add to 50 µl with nf H20; 60`@ 37°C)
Ligation of:
1. K228000 + RBS
2. K322127 + supD-tRNA
3. lacZ + T7-Promotor
08-07-2011
Results
Cloning
preparative gel for ligation from 07-07-11; 1% TAE, 8 wells, 120 V, 400 mA, 90';
30 µl DNA + 6 µl loading dye (6x) -> 36 µl, 35 µl loaded on gel
7,5 µl 2-log DNA ladder
11-07-2011
Cloning
Ethanol precipitation (1/10 of volume 3M NaAc (pH 5.8), 2.5 X EtOH (100%, icecold)) of:
- Backbone (T7-Promotorpart, cut with S, P): 200 µl
- RBS (cut with S, P): 70 µl
- K228000 (cut with X, P): 90 µl
- K322127 (PCR-product redlight w/o lacZ; cut with E, S): 180 µl
- lacZ (cut with X, P): 90 µl
Add NaAc and EtOH (100%), invert 5x, incubate 30' @ -80°C
centrifuge 30' @ 4°C, 10000 rcf
no visible pellet -> centrifuge again, 30' @ 4°C, 18000 rcf
discard supernatant
wash with EtOH (70%, icecold) -> centrifuge 5' @ 4°C, 18000 rcf
discard supernatant
dry pellet (30' @ RT)
resuspend in 10 µl nuclease-free H20 (at no time visible pellet. nanodrop measurements suggested no detectable amount of DNA)
Restriction
Restriction digest (total volume: 50 µl; Enzymes: 1 µl each; 5 µl NEB 4 buffer (10x); incubation: 1 h @ 37°C; inactivation: 20 ' @ 80 °C) of PCR samples 2 and 4 (HF-buffer) from 05-07-11:
DNA: 10 µl (sample 2: 1 µl primer [10 µM], -DMSO; sample 4: 1 µl primer [10 µM], +DMSO)
H20: 33 µl
Enzymes: E, S
Preparative agarosegel (1%, 1xTAE, 8 wells, 120 V, 400 mA, 90'):
35 µl restriction digest + 7 µl loading dye (6x) -> 42 µl; 40 µl loaded on gel
7 µl 2-log DNA ladder
lanes:
1.) 2-log
2.) PCR-sample 2 from 05-07-11
3.) PCR-sample 4 from 05-07-11
12-07-2011
Cloning
Digest
Restriction digest (total volume: 50 µl; DNA: 1,5 µg; Enzyme: 1 µl each; 5 µl NEB 4 buffer (10x); incubation: 1 h @ 37°C; inactivation: 20 ' @ 80 °C) of the following parts:
- lacZ: 94 ng/µl -> 16 µl; H20: 27 µl; Enzymes: X, P
- T7-Promotor: 106,6 ng/µl -> 14 µl; H20: 29 µl; Enzymes: S, P
- SupD-tRNA: 150 ng/µl -> 10 µl; H20: 33 µl; Enzymes: E, X (#1, #2: double preparation for validation of different subsequent DNA purification steps)
- K238013 (blue light promotor): 233 ng/µl -> 6,5 µl; H20: 36,5 µl; Enzymes: E, X
- B0015 (Term.): 141 ng/µl -> 10,5 µl; H20: 32,5 µl; Enzymes: E, S
- RBS: 108 ng/µl -> 14 µl; H20: 29 µl; Enzymes: S, P
preparative agarosegel (1%, 1xTAE, 8 wells, 120 V, 400 mA, 90'):
35 µl restriction digest + 7 µl loading dye (6x) -> 42 µl; 40 µl loaded on gel
7 µl 2-log DNA ladder
1) DNA ladder
2) lacZ
3) T7 Promotor
4) supD tRNA
5) K328013
6) RBS
preparative agarosegel (2%, 1xTAE, 8 wells, 120 V, 400 mA, 90'):
35 µl restriction digest + 7 µl loading dye (6x) -> 42 µl; 40 µl loaded on gel
7 µl 2-log DNA ladder
0) DNA ladder (pipette tip fell off)
1) DNA ladder
2) B0015 (Term.)
Ligation
Ligation (total volume: 30 µl; 10 µl of each DNA sample; 3 µl T4-ligase buffer (10x); 1 µl Quick-Ligase; 6 µl nf H20; 30`@ 37°C)
Ligation of:
1. lacZ + T7-Promotor
2. K322127 (PCR-Product) + supD-tRNA
3. K238013 + B0015
13-07-2011
Cloning
MiniPrep
Preparation of 24 overnight-cultures (5 ml each) for miniprep
Picking
picking of 3 clones (labelled a, b and c) per positive (= visible colonies) plate:
plate 1: lacZ + T7-promotor AFTER purification -> no colonies visible; Kan-resistance
plate 2: lacZ + T7-promotor BEFORE purification -> colonies; Kan-resistance
plate 3: PCR-product (from K322127) + supD-tRNA AFTER purification -> colonies; Amp-resistance
plate 4: PCR-product (from K322127) + supD-tRNA BEFORE purification -> colonies; Amp-resistance
plate 5: K238013 + B0015 AFTER purification -> colonies; Amp-resistance
plate 6: K238013 + B0015 BEFORE purification -> colonies (half normal, half slightly red); Amp-resistance
plate 7: K228000 + RBS AFTER purification -> colonies; Amp-resistance
plate 8: K228000 + RBS BEFORE purification -> colonies; Amp-resistance
plate 9: PCR-product (from K322127) + supD-tRNA BEFORE purification [ligated with T4 ligase] -> colonies; Amp-resistance
picking occured with 200 µl pipette tip, thrown into 15 ml Falcon with 5 ml LB + antibiotic (1:1000)
shaking started at 18:00 p.m., 200 rpm, 37°C, o/n
Other Work
Analytical agarosegel [130711-ligation-control.tif] (1%, 1xTBE, 20 wells, 120 V, 400 mA, 90') for control of ligations from 12-07-11:
10 µl ligation sample + 2 µl loading dye (6x) -> 12 µl, 10 µl loaded on gel
5 µl 2-log DNA ladder
lanes:
1) 2-log
2) lacZ + T7-promotor AFTER purification
3) lacZ + T7-promotor BEFORE purification
4) PCR-product (from K322127) + supD-tRNA AFTER purification
5) PCR-product (from K322127) + supD-tRNA BEFORE purification
6) K238013 + B0015 AFTER purification
7) K238013 + B0015 BEFORE purification
8) K228000 + RBS AFTER purification
9) K228000 + RBS BEFORE purification
10) PCR-product (from K322127) + supD-tRNA BEFORE purification [ligated with T4 ligase]
11) 2-log
14-07-2011
Results
Ligation worked for K228000 + RBS (all six samples 7 a-c and 8 a-c), K238013 + B0015 (6c), lacZ+Z7-Promotor (2b)
Ligation failed for K322127 + supD-tRNA
Cloning
MiniPrep
- miniprep (4 ml) of the 24 over-night cultures from 13-7-11:
2a-c: lacZ + T7-promotor BEFORE purification
3a-c: PCR-product (from K322127) + supD-tRNA AFTER purification
4a-c: PCR-product (from K322127) + supD-tRNA BEFORE purification
5a-c: K238013 + B0015 AFTER purification
6a-c: K238013 + B0015 BEFORE purification
7a-c: K228000 + RBS AFTER purification
8a-c: K228000 + RBS BEFORE purification
9a-c: PCR-product (from K322127) + supD-tRNA BEFORE purification [ligated with T4 ligase]
- measurement of the DNA concentration with nanodrop
2a: 25 ng/µl
2b: 215 ng/µl
2c: 9.5 ng/µl
3a: 85 ng/µl
3b: 53.5 ng/µl
3c: -
4a: 226 ng/µl
4b: 20 ng/µl
4c: 57.5 ng/µl
5a: 68.5 ng/µl
5b: -
5c: 20.5 ng/µl
6a: 360 ng/µl
6b: 143 ng/µl
6c: 305 ng/µl
7a: 443 ng/µl
7b: 218 ng/µl
7c: 204 ng/µl
8a: 228 ng/µl
8b: 268 ng/µl
8c: 218 ng/µl
9a: 38.5 ng/µl
9b: 16 ng/µl
9c: 16.5 ng/µl
Restriction
restriction digest of 19 plasmids from the miniprep (without 2c, 3c, 5b, 9b, 9c)
Restriction digest (total volume: 30 µl; DNA: 0,5µg; Enzyme (EcoR1 + Psi): 1 µl each; 3 µl NEB 4 buffer (10x); incubation: 50´@ 37°C)
Analytical agarosegel (1%, 1xTBE, 20 wells, 120 V, 400 mA, 50') for control of transformation success from 13-07-11:
10 µl sample + 2 µl loading dye (6x) -> 12 µl, 11 µl loaded on gel
5 µl 2-log DNA ladder
lanes:
0.) 2 log
1.) 2a -> lacZ + T7-promotor BEFORE purification
2.) 2b -> lacZ + T7-promotor BEFORE purification
3.) 3a -> PCR-product (from K322127) + supD-tRNA AFTER purification
4.) 3b -> PCR-product (from K322127) + supD-tRNA AFTER purification
5.) 4a -> PCR-product (from K322127) + supD-tRNA BEFORE purification
6.) 4b -> PCR-product (from K322127) + supD-tRNA BEFORE purification
7.) 4c -> PCR-product (from K322127) + supD-tRNA BEFORE purification
8.) 5a -> K238013 + B0015 AFTER purification
9.) 5c -> K238013 + B0015 AFTER purification
10.) 6a -> K238013 + B0015 BEFORE purification
11.) 6b -> K238013 + B0015 BEFORE purification
12.) 6c -> K238013 + B0015 BEFORE purification
13.) 7b -> K228000 + RBS AFTER purification
14.) 7a -> K228000 + RBS AFTER purification
15.) 7c -> K228000 + RBS AFTER purification
16.) 8a -> K228000 + RBS BEFORE purification
17.) 8b -> K228000 + RBS BEFORE purification
19.) 8c -> K228000 + RBS BEFORE purification
19.) 9a -> PCR-product (from K322127) + supD-tRNA BEFORE purification [ligated with T4 ligase]
15-07-2011
Results
in all samples and plates failed the experiment. There was no useable DNA detected
Cloning
PCR
Colony PCR for the ligation of K322127 and supD-tRNA. Therefore 15 colonies of the plates 3,4 and 9 (from 13-7-11) were resuspended in 50 µl nuclease free water. The experimet failed
Analytical agarosegel (1%, 1xTBE, 20 wells, 120 V, 400 mA, 90') for control of lthe colony-PCR from 15-7-11: 10 µl ligation sample + 2 µl loading dye (6x) -> 12 µl, 10 µl loaded on gel 5 µl 2-log DNA ladder
plate 3:
lanes:
1. 2-log
2. positive control
3-17. colonies from plate 3
18. positive control
plate 4:
lanes:
1. 2-log
2. negative control
3-17. samples from the colonies of plate 4
plate 9:
lanes:
1. 2-log
2-17. samples from colonies of plate 9
19-07-2011
Results
restriction digest worked
Cloning
Restriction
Restriction digest of K238013+B0015 with S, P and K228000 (T7-Polymerase) + RBS with X, P
Restriction digest (total volume: 30 µl; DNA: 1 µg; Enzyme 1 µl each; 3 µl NEB 4 buffer (10x); incubation: 1 h @ 37°C)
The used DNA-templates were 8a (14-7-11) and 6c (14-7-11)
preparative agarosegel (1%, 1xTAE, 8 wells, 120 V, 400 mA, 90'):
30 µl restriction digest + 6 µl loading dye (6x) -> 35 µl loaded on gel
lanes:
1.) 2 log- DNA ladder 7 µl
2.) K22800 + RBS
3.) K238013 + B0015
4.) K322127 from an earlier restriction digest
-> DNA extraction from gel via mi-Gel-extraction kit from metabiom
20-07-2011
results
ligation success inconclusive
Cloning
Ligation
ligation of K228000-RBS with K238013-B0015 and K322127 with subD-tRNA.
Ligation (total volume: 30 µl; 10 µl of each DNA sample; 3 µl T4-ligase buffer (10x); 1 µl Quick-Ligase; 6 µl nf H20; 30`@ 37°C)
Transformation
transformation (40 µl competent cells; 1 µl DNA; Electroporation at 1510 V; incubation in Soc-Medium for 1 h @ 37°C, 50 µl on Amp-agar plate)
analytical agarosegel (1%, 1xTBE, 8 wells, 120 V, 400 mA, 90')
10 µl DNA + 2 µl loading dye -> 10 µl on gel
Lanes:
1.) 7 µl 2-log DNA ladder
2.) K228000-RBS + K238013 - B0015 (upper band in preparative gel)
3.) K228000-RBS + K238013 - B0015 (lower band in preparative gel)
4.) K322127-tRNA
21-07-2011
Cloning
Restriction
control restriction digest with the Enzymes E, P of 2b (=ligation product lacZ+T7 promotor), 6c (=ligation product K238013+B0015) and 8a (ligation product K228000+RBS).
Restriction digest (total volume: 30 µl; DNA: 0.5 µg; Enzyme 1 µl each (E, P); 3 µl NEB 4 buffer (10x); incubation: 1 h @ 37°C)
analytical gelelektrophoresis (1%, 1x TBE, 12 wells, 120 V, 400 mA, 90´)
lanes:
1.) 2b ( =ligation product lacZ+T7-Promotor)
3.) 6c (=ligation product K238013 B B0015)
5.) 8A (=ligation product K228000 + RBS)
6.) 2-log DNA ladder
Other Work
production of LB-Agar plates with ampicillin
- 5 ml overnight culture of the transformated cells from 20-7-11. From each transformation 3 colonies were picked and cultivated in 5 ml LB-medium with Ampicillin.
-> 10 a-c: ligation product K322127 + tRNA
-> 11 a-c: ligation product K228000-RBS + K238013+B0015
- 5ml overnight culture of 218 (pBADlacZ) with Ampicillin
- 5 ml overnight culture of BM28 with Kanamycin
-> both cultures are used for gelrite experiments
22-07-2011
Cloning
MiniPrep
miniprep of the 4 ml overnight cultures from 21-7-11 with metabion
10 a-c: ligation product K322127 + tRNA
11 a-c: ligation product K228000-RBS + K238013+B0015
- nanodrop measurement of the DNA concentration:
10 a: 62 ng/µl
10 b: 42 ng/µl
10 c: 38.5 ng/µl
11 a: 51 ng/µl
11 b: 47.5 ng/µl
11 c: 47.5 ng/µl
Restriction
control regristiction digest of 10 a-c (ligation product K322127 + tRNA) and 11 a-c (ligation product K228000-RBS + K238013+B0015) with E, P
Restriction digest (total volume: 30 µl; DNA: 0.3 µg; Enzyme 1 µl each (E, P); 3 µl NEB 4 buffer (10x); incubation: 1 h 10` @ 37°C, inactivation 20´@ 80°C)
analytical gelelectrophoresis (1%, TAE 1x, 12 wells, 120 V, 400 mA, 120`)
lanes:
1.) 2-log ( 1 µl 2-log, 1 µl loading buffer (6x), 10 µl water) -> 10 µl loaded on gel
2.) 10 a
3.) 10 b
4.) 10 c
5.) 11 a
6.) 11 b
7.) 11 c
 "
"