Team:Wageningen UR/Project/ModelingProj2
From 2011.igem.org
(→Modeling: Fungal Track 'n Trace) |
(→Modeling: Fungal Track 'n Trace) |
||
| (One intermediate revision not shown) | |||
| Line 3: | Line 3: | ||
<style type="text/css"> | <style type="text/css"> | ||
| - | ul li a. | + | ul li a.currentlinkfungus2m { |
color: black !important; | color: black !important; | ||
} | } | ||
| Line 9: | Line 9: | ||
ul li a.currentlinktop3 { | ul li a.currentlinktop3 { | ||
color: #63a015 !important; | color: #63a015 !important; | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
} | } | ||
| Line 30: | Line 25: | ||
[[File:Spee_FTnT_Model-1_WUR.jpg|600px|center]] | [[File:Spee_FTnT_Model-1_WUR.jpg|600px|center]] | ||
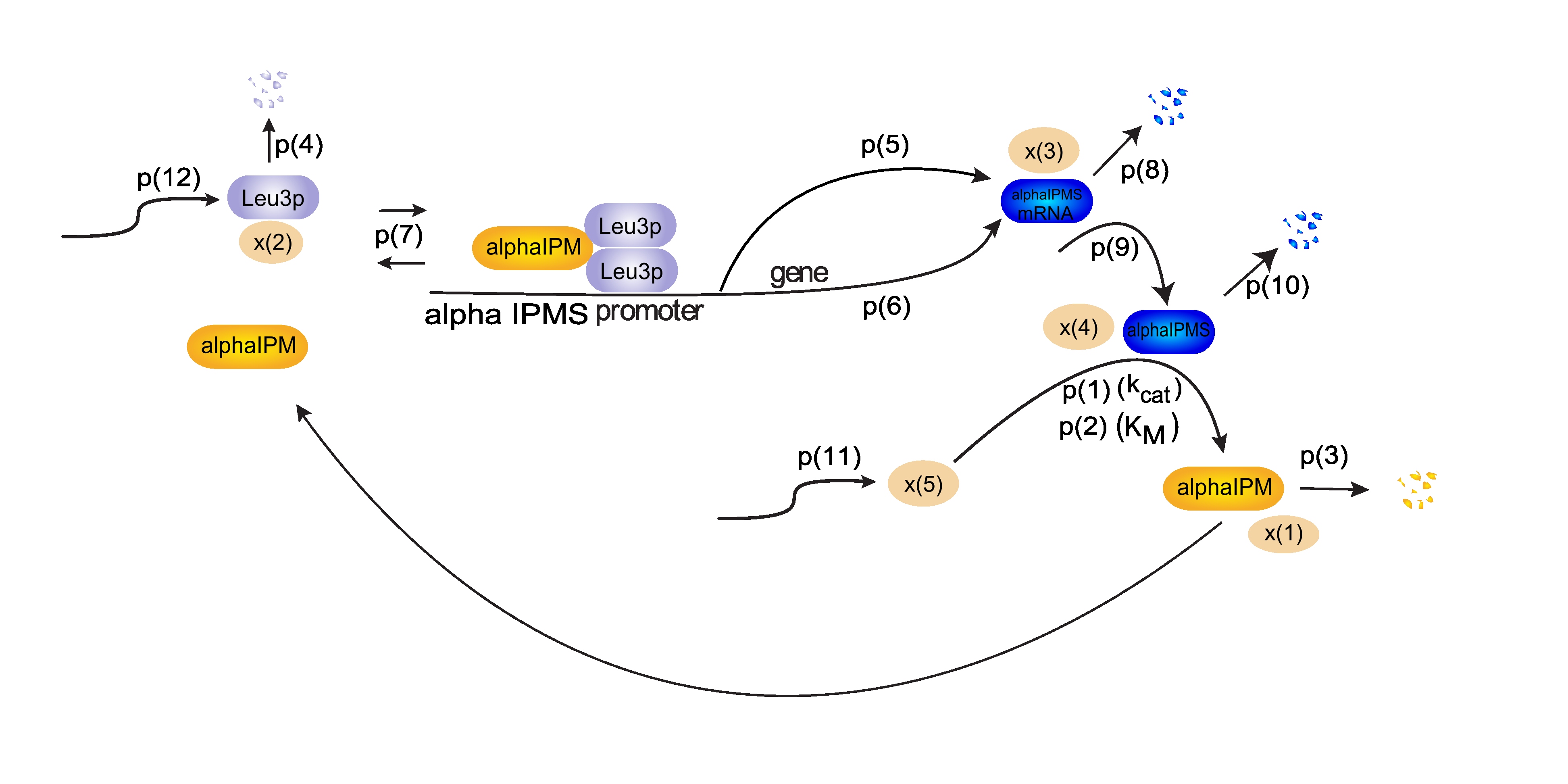
| - | The Leu3p (x(2)) dimer blocks transcription from the alpha-IPM synthase (alpha IPMS) promoter. Alpha-IPM (x(1)) binding to this dimer releases it from the promoter and enhances the transcription rate. p(5) and p(6) model both constitutive expression and the transcription rate increase upon alpha-IPM binding. From alpha-IPMS mRNA (x(3)) the protein is formed, modelled by p(9). Alpha-IPMS (x(4)) enzymatically produces alpha-IPM, which is then incorporated in the beginning of this loop again. Intercellular transport of alpha-IPM creates a signal transduction through a | + | The Leu3p (x(2)) dimer blocks transcription from the alpha-IPM synthase (alpha IPMS) promoter. Alpha-IPM (x(1)) binding to this dimer releases it from the promoter and enhances the transcription rate. p(5) and p(6) model both constitutive expression and the transcription rate increase upon alpha-IPM binding. From alpha-IPMS mRNA (x(3)) the protein is formed, modelled by p(9). Alpha-IPMS (x(4)) enzymatically produces alpha-IPM, which is then incorporated in the beginning of this loop again. Intercellular transport of alpha-IPM creates a signal transduction through a hypha. |
Additionally, x(5) is required for modelling the enzymatic reaction. Its formation is described by p(11). Each type of molecule has also been assigned a degradation constant, such as p(8) for x(3). | Additionally, x(5) is required for modelling the enzymatic reaction. Its formation is described by p(11). Each type of molecule has also been assigned a degradation constant, such as p(8) for x(3). | ||
}} | }} | ||
Latest revision as of 20:20, 21 September 2011
 "
"