|
|
| (4 intermediate revisions not shown) |
| Line 9: |
Line 9: |
| | **sent to Genewiz to be sequenced with His3_F primer | | **sent to Genewiz to be sequenced with His3_F primer |
| | | | |
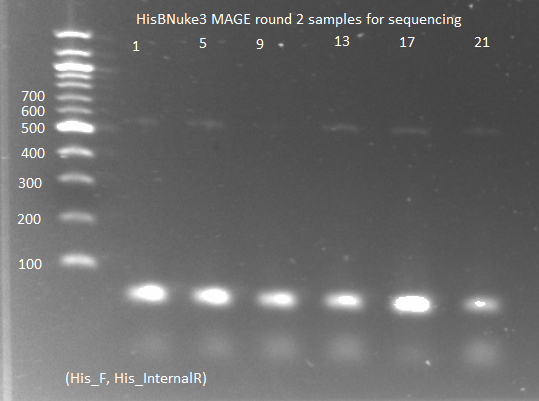
| - | [[File: 2011.07.01.hisBnuke3MAGE2_for_seq2(labeled).png|thumb|none|HisBNuke3 MAGE round 2 samples for sequencing 7/1/11]] | + | [[File: HARV2011.07.01.hisBnuke3MAGE2_for_seq2(labeled).png|thumb|none|HisBNuke3 MAGE round 2 samples for sequencing 7/1/11]] |
| | | | |
| | '''Lambda Red recombination results:''' | | '''Lambda Red recombination results:''' |
| Line 21: |
Line 21: |
| | **157.4ng/µL 260/280=1.9; 129.2ng/µL 260/280=1.88 | | **157.4ng/µL 260/280=1.9; 129.2ng/µL 260/280=1.88 |
| | | | |
| - | [[File: 2011.07.01.pZE21Gbackbonesuccess(labeled).png|thumb|none|pZE21G backbone 7/1/11]] | + | [[File: HARV2011.07.01.pZE21Gbackbonesuccess(labeled).png|thumb|none|pZE21G backbone 7/1/11]] |
| | | | |
| | '''Selection system plates:''' | | '''Selection system plates:''' |
| - | *Made 3-AT and 5-FOA plates following 2006 Meng Nature Protocol (see [[Protocols#Selection Plates|protocols]] for details). 5-FOA plates have a blue stripe, 3-AT plates (0.5 mL 3-AT) have a green stripe. | + | *Made 3-AT and 5-FOA plates following 2006 Meng Nature Protocol (see [https://2011.igem.org/Team:Harvard/Protocols protocols] for details). 5-FOA plates have a blue stripe, 3-AT plates (0.5 mL 3-AT) have a green stripe.</div> |
| - | </div> | + | <div id="704" style="display:none"> |
| | | | |
| - |
| |
| - | <div id="704" style="display:none">
| |
| | ==July 4th== | | ==July 4th== |
| - | | + | FIREWORKS</div> |
| - | FIREWORKS | + | |
| - | </div> | + | |
| - | | + | |
| - | | + | |
| | <div id="705" style="display:none"> | | <div id="705" style="display:none"> |
| | ==July 5th== | | ==July 5th== |
| Line 63: |
Line 57: |
| | **Because of KAPA shortage, only ran samples 6-8 from 100µL 6/29 plate and 6-7 from 2mL 6/29 plate plus (ΔHisB)ΔpyrFΔrpoZ+pKD46 from 6/29 culture as a control (should produce a band around 300bp, while the insert would make the band 2-3kb) | | **Because of KAPA shortage, only ran samples 6-8 from 100µL 6/29 plate and 6-7 from 2mL 6/29 plate plus (ΔHisB)ΔpyrFΔrpoZ+pKD46 from 6/29 culture as a control (should produce a band around 300bp, while the insert would make the band 2-3kb) |
| | **Ran on E-Gel with team TolC's samples, but there was an issue loading the gel, so also ran separately on another gel. Unfortunately most of the PCR reactions strangely did not work, but two of the samples appeared not to have the insert (see below). | | **Ran on E-Gel with team TolC's samples, but there was an issue loading the gel, so also ran separately on another gel. Unfortunately most of the PCR reactions strangely did not work, but two of the samples appeared not to have the insert (see below). |
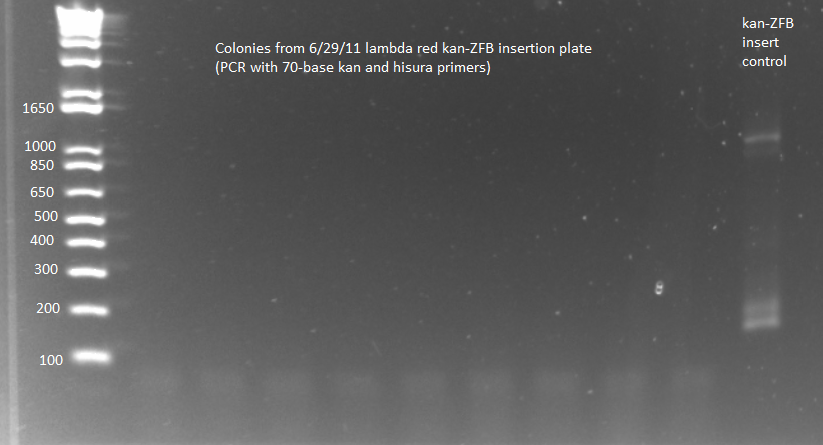
| - | [[File:2011.07.05.kanZFBwp_tolC+kanZFBinsert_check.png|200px|thumb|none|2011.07.05.kanZFBwp_tolC+kanZFBinsert_check.ai]] | + | [[File:HARV2011.07.05.kanZFBwp_tolC+kanZFBinsert_check.png|200px|thumb|none|2011.07.05.kanZFBwp_tolC+kanZFBinsert_check.ai]] |
| | | | |
| | | | |
| Line 105: |
Line 99: |
| | *Teams: Make sure everything is organized in your dropbox! | | *Teams: Make sure everything is organized in your dropbox! |
| | *''*Announcing a New (and Final) Plasmid Name*'' '''PSR01''': formerly known as Wolfe Plasmid/Vatsan's Plasmid/Selection | | *''*Announcing a New (and Final) Plasmid Name*'' '''PSR01''': formerly known as Wolfe Plasmid/Vatsan's Plasmid/Selection |
| - | **Contains ~ - KAN - ZFbs - weak promoter - URA3 - HIS3- ~ | + | **Contains ~ - KAN - ZFbs - weak promoter - URA3 - HIS3- ~</div> |
| - | </div> | + | |
| - | | + | |
| - | | + | |
| | <div id="706" style="display:none"> | | <div id="706" style="display:none"> |
| | ==July 6th== | | ==July 6th== |
| Line 121: |
Line 112: |
| | **(ΔHisB)ΔpyrFΔrpoZ+pKD46 from 6/29 culture used as a control | | **(ΔHisB)ΔpyrFΔrpoZ+pKD46 from 6/29 culture used as a control |
| | | | |
| - | [[File: 2011.07.06.kanZFBinsertcheck1(labeled).png|thumb|none|1529620 locus PCR 1 7/6/11]] | + | [[File: HARV2011.07.06.kanZFBinsertcheck1(labeled).png|thumb|none|1529620 locus PCR 1 7/6/11]] |
| - | [[File: 2011.07.06.kanZFBinsertcheck2(labeled).png|thumb|none|1529620 locus PCR 2 7/6/11]] | + | [[File: HARV2011.07.06.kanZFBinsertcheck2(labeled).png|thumb|none|1529620 locus PCR 2 7/6/11]] |
| | | | |
| | *NM media has 1% of the glucose concentration written in the protocol, it is 0.4mg/ml instead of 40mg/ml. Nevertheless, our overnight culture of pKD46 grew successfully so we will continue to use the media unless other problems arise. | | *NM media has 1% of the glucose concentration written in the protocol, it is 0.4mg/ml instead of 40mg/ml. Nevertheless, our overnight culture of pKD46 grew successfully so we will continue to use the media unless other problems arise. |
| Line 131: |
Line 122: |
| | **results from 2 E Gels: PCR not very successful. Colonies showed no bands while the control showed only a side product too small to be the full insert. | | **results from 2 E Gels: PCR not very successful. Colonies showed no bands while the control showed only a side product too small to be the full insert. |
| | | | |
| - | [[File: 2011.07.06.kanZFBinsertcheck_longprimer1(labeled).png|thumb|none|Kan-ZFB check using kan and hisura primers 7/6/11]] | + | [[File: HARV2011.07.06.kanZFBinsertcheck_longprimer1(labeled).png|thumb|none|Kan-ZFB check using kan and hisura primers 7/6/11]] |
| | | | |
| | *Grew up 24 more kan-ZFB lambda red recombined colonies (12 from 100µL 6/29 plate, 12 from 2mL 6/29 plate) in LB+kan in 96 well plate for PCR tomorrow | | *Grew up 24 more kan-ZFB lambda red recombined colonies (12 from 100µL 6/29 plate, 12 from 2mL 6/29 plate) in LB+kan in 96 well plate for PCR tomorrow |
| Line 156: |
Line 147: |
| | **4°C forever | | **4°C forever |
| | | | |
| - | [[File:KAN-ZFB-WP2 Initial Attempt.png|200px|thumb|none|KAN-ZFB-WP]] | + | [[File:HARVKAN-ZFB-WP2 Initial Attempt.png|200px|thumb|none|KAN-ZFB-WP]] |
| | | | |
| | ===Web Design=== | | ===Web Design=== |
| | *Project description | | *Project description |
| - | *Web design brainstorming;exploring past designs and templates | + | *Web design brainstorming;exploring past designs and templates</div> |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | <div id="707" style="display:none">
| + | |
| - | ==July 7th==
| + | |
| - | | + | |
| - | ===Team ZF Assembly===
| + | |
| - | Our primers still haven't arrived. We are working with the Web Design Team.
| + | |
| - | | + | |
| - | ===Team Wolfe Selection===
| + | |
| - | | + | |
| - | *PCR for Round 2 MAGE cultures in LB media
| + | |
| - | **The samples came from the overnight culture of MAGE/control strains grown in LB+amp, NM+his, and NM. Oddly, the (∆HisB)∆pyrF∆rpoZ+pKD46 control grew only in NM+His and LB, and not in plain NM.
| + | |
| - | ** Samples
| + | |
| - | *** Control= 1 ss+pKD46
| + | |
| - | *** 11 cultures from 1μL plate in LB media
| + | |
| - | *** 12 cultures from 10μL plate in LB media
| + | |
| - | **Program
| + | |
| - | ***Anneal at 56°
| + | |
| - | ***Elongation for 90 seconds
| + | |
| - | ***All else according to standard KAPA protocol
| + | |
| - | **E gel of control and 1µL plate colonies 1-8 to check PCR worked (it was successful)
| + | |
| - | **samples prepared for sequencing tomorrow
| + | |
| - | | + | |
| - | [[File: 2011.07.07.hisBseq labeled.png|thumb|none|MAGE 2 samples for sequencing 7/7/11]]
| + | |
| - | | + | |
| - | *Kan-ZFB insert check PCR results:
| + | |
| - | **the overnight cultures grown yesterday were used in a PCR (same KAPA protocol as before) around the 1529620 locus. Not all the cultures grew; 100µL plate cultures 1-5 and 2mL plate cultures 1-12 were used.
| + | |
| - | **Results: some of the reactions, strangely, did not work, but the ones that did showed a short band and thus did not have the insert (see below)
| + | |
| - | | + | |
| - | [[File: 2011.07.07,kanZFBinsert_check1 labeled.png|thumb|none|1529620 locus PCR 7/7/11]]
| + | |
| - | [[File: 2011.07.07,kanZFBinsert_check2 labeled.png|thumb|none|1529620 locus PCR 7/7/11]]
| + | |
| - | | + | |
| - | *HisBNuke3 MAGE round 3:
| + | |
| - | **used same protocol as previously described, using 1.5mL mid-log culture from MAGE round 2
| + | |
| - | **will recover overnight so that we can reinoculate and continue MAGE rounds tomorrow
| + | |
| - | | + | |
| - | *NM medium:
| + | |
| - | **added 3.9g glucose to 100mL NM medium to bring the glucose concentration up to the correct level.
| + | |
| - | **Because (∆HisB)∆pyrF∆rpoZ+pKD46 did not grow in plain NM, we are curious to see if that was due to low glucose levels. One colony from a plate was put in 3mL NM(gluc) and will be grown overnight.
| + | |
| - | | + | |
| - | *HisB MAGE2:
| + | |
| - | **In case the sequencing does not show successful MAGE, we will grow overnight 48 more colonies (24 from each plate) in 100µL of NM(gluc) and LB+amp
| + | |
| - | | + | |
| - | ===Team TolC===
| + | |
| - | *making culture tubes of ECNR2 and ECNR2delTolC for glycerol stock (no Amp)
| + | |
| - | *making culture tubes of ECNR2 for MAGE to delete rpoZ (one with amp, one without)
| + | |
| - | *dilutions of rpoZ deletion MAGE oligo, 20µM
| + | |
| - | *dilution of rpoZ deletion MAGE oligo, 1µM, for MAGE.
| + | |
| - | *serial dilution of ECNR2 after MAGE round 1, and round 2, with 20µl of 100x dilution of concentrated culture. For plating onto amp plates.
| + | |
| - | *During Gel extraction, ignored 3x volume in step 2 and instead used 500 µl of QG buffer. Additionally, used 18 µl of EB buffer and not water in step 13
| + | |
| - | | + | |
| - | | + | |
| - | [[File:KAN-ZFB-WP.png|200px|thumb|none|Kan-ZFB-WP]]
| + | |
| - | | + | |
| - | ===Web Design===
| + | |
| - | ====Brainstorming====
| + | |
| - | *Minimalist toolbar, metallic crimson color
| + | |
| - | *"Popout" menu below, ''not'' dropdown
| + | |
| - | *Slideshow on front page
| + | |
| - | **First slide: Brief text project description, brief youtube video description (in layman's terms)
| + | |
| - | *See the [[Wiki|Public Wiki]] page for our ideas about the pages we should include, page set up, and buttons
| + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | <div id="708" style="display:none">
| + | |
| - | ==July 8th==
| + | |
| - | | + | |
| - | ===Team ZF Assembly===
| + | |
| - | ====PCR of omega+zif268 with overhangs====
| + | |
| - | We performed a PCR to take out our omega subunit plus zif268. We used the following recipe:
| + | |
| - | *10 ul KAPA readymix
| + | |
| - | *0.75 ul of hindIII (primer)
| + | |
| - | *0.75 ul of wF+plLacO (primer)
| + | |
| - | *1.2 ul template
| + | |
| - | *12.3 ul water
| + | |
| - | | + | |
| - | The first steps of the PCR involved heating at 95* (for 5 minutes) and 98* (for 20 seconds). Annealing temperature was 56*. Extension time was 30 seconds.
| + | |
| - | | + | |
| - | The product of the PCR should be 709 bps. It includes the omega subunit, zif268, as well as overlap sequences homologous to the selection plasmid.
| + | |
| - | We ran an e-gel to ensure that our desired product was present:
| + | |
| - | | + | |
| - | ====Gel purification of PCR product====
| + | |
| - | We had to gel purify our PCR product in preparation for isothermal assembly, using [[Protocols#Gel_purification| our protocol for gel purification]]. The result from running the gel can be seen below - we cut out the bands at 709 bp, the expected size of our product.
| + | |
| - | [[Image:2011.07.08_omega_+_zif268_with_overhangs_annotated.png]]
| + | |
| - | | + | |
| - | In the final step of the purification process, we eluted our DNA in buffer EB. We used the nanodrop spectophotometer to determine the concentration of DNA present.
| + | |
| - | *The concentration was 54.3 ng/ul.
| + | |
| - | | + | |
| - | ===Team Wolfe Selection===
| + | |
| - | '''MAGE round 3:'''
| + | |
| - | *MAGE cultures that recovered overnight were saturated, so we made 1:1000, 1:10000, and 1:100000 dilutions and plated 20µL of each onto amp plates and left at 30˚C overnight.
| + | |
| - | | + | |
| - | '''MAGE round 2 cultures from yesterday:'''
| + | |
| - | *We did a PCR (His_F and and His_R primers) on 24 of the 48 cultures we grew and sent them off for sequencing.
| + | |
| - | | + | |
| - | '''Results of NM culture:'''
| + | |
| - | *The selection strain (∆HisB∆pyrF∆rpoZ+pKD46) did not grow in NM, which is worrisome. None of the MAGE samples grew in NM either, so either they all have HisB knocked out or there is something wrong already with the strain's His pathway.
| + | |
| - | *We will grow up selection strain (without pKD46) from glycerol stock in LB+tet and transform with Vatsan's plasmid tomorrow to see if that has any effect on the his-deficiency phenotype.
| + | |
| - | | + | |
| - | '''Kan-ZFB insertion: modified nucleotides:'''
| + | |
| - | *Primers finally arrived that would put modified nucleotides on the 5' ends of the insert, which should improve lambda red efficiency.
| + | |
| - | *PCR: KAPA protocol with Kan forward phosphorthioate and URA2 reverse phosphorthioate, 56˚C annealing, 90 sec elongation, 25 cycles, 1µL kan-ZFB-hisura DNA as template
| + | |
| - | *Results: PCR only produced a smaller side product. This is strange since we also ran a gel of all our different kan-ZFB-hisura purifications and they all run at the correct size (see images below).
| + | |
| - | | + | |
| - | [[File: 2011.07.08.ECnr2MAGEround2-21to24delrpoZ&thioKAN-ZFB labeled.png|thumb|none|Thio kan-ZFB 7/8/11]]
| + | |
| - | [[File: 2011.07.08.kanzfbDNA(labeled).png|thumb|none|kan-ZFB insert purifications]]
| + | |
| - | | + | |
| - | ===Team TolC===
| + | |
| - | *Performed re-inoculation of ECNR2-Placed into tube labeled ECNR 2 MAGE 3 in 30 degree shaker.
| + | |
| - | *grew up 24 colonies total for sequencing ECNR2 for rpoZ deletion (the colonies appear to have grown much faster than expected).
| + | |
| - | **rows A (MAGE round 1) and B (MAGE round 2), for PCR and sequencing. to be placed in the 4C refrigerator after use as template.
| + | |
| - | *RPOZ PCR-Machine 6
| + | |
| - | **Primer Annealing = 56 degrees
| + | |
| - | **Extension 48 = seconds
| + | |
| - | | + | |
| - | ===Team Web Design===
| + | |
| - | *Edited/Re-drafted our project description, see Dropbox (It's a bit long as is now, but the excess informaiton that we cut out can still probably be used somewhere on our website
| + | |
| - | *Sketched draft pages of the public wiki (Please refer to the [[Wiki|Public Wiki]] page)
| + | |
| - | **Homepage- first draft sketch done based on the characteristics and requirements we'd discussed. However, there are a few issues (i.e., how to include some of the iGEM wiki reuqirements); please see these listed on the [[Wiki|Public Wiki]] page.
| + | |
| - | </div>
| + | |
| - | | + | |
| - | <div id="709" style="dispaly:none">
| + | |
| - | ==July 9==
| + | |
| - | ===Team ZF===
| + | |
| - | ====Isothermal Assembly====
| + | |
| - | We performed an isothermal assembly in order to insert omega+zif268 into the plasmid containing the spec backbone.
| + | |
| - | *The concentration of the plasmid (previously prepared) was 157.4 ng/ul.
| + | |
| - | *The concentration of the w+zif268 was 54.3 ng/ul.
| + | |
| - | | + | |
| - | Using [[Protocols#Isothermal_assembly|our protocols]] we determined we needed:
| + | |
| - | *0.63 ul backbone (length: ~2.2 kb)
| + | |
| - | *0.59 ul w+zif268 (length: ~0.7 kb)
| + | |
| - | *2.78 ul water
| + | |
| - | *15.0 ul isothermal assembly solution
| + | |
| - | | + | |
| - | ====Transformation of electro-competent ''E. coli''====
| + | |
| - | We electroporated our cells so they would take up the plasmids, using:
| + | |
| - | *1.0 ul DNA
| + | |
| - | *40.0 ul cells
| + | |
| - | *500 ul LB
| + | |
| - | | + | |
| - | We plated the cells, [[Protocols#Cultures|using this protocol]], on spec plates, one of which contained 50 ul of cells and the other 150 ul of cells. The cells have been incubated at 37*, and will be left there overnight. Cells that have taken up the plasmid have spec-resistance and should be able to survive. Tomorrow, we will be picking colonies and performing PCR on them to determine whether they contain w+zif268.
| + | |
| - | | + | |
| - | ===Team Wolfe===
| + | |
| - | *MAGE3 plates look good--we'll choose colonies for sequencing on Monday.
| + | |
| - | *Transformation of selection strain with Vatsan's plasmid:
| + | |
| - | **We're curious to see whether Vatsan's plasmid (Zif268 omega, ZFB, wp, hisura) will have an effect on the (∆HisB)∆pyrF∆rpoZ strain's growth despite the seemingly intact HisB.
| + | |
| - | **Transformed mid-log selection strain (reinoculated from overnight culture) with about 100ng of Vatsan's plasmid following standard procedure.
| + | |
| - | **After 2 hr recovery plated on amp plates with 100µL or 1mL
| + | |
| - | </div>
| + | |
| - | | + | |
| - | <div id="710" style="display:none">
| + | |
| - | ==July 10th==
| + | |
| - | ===Team ZF===
| + | |
| - | ====PCR to check for omega+zif268====
| + | |
| - | All colonies that grew on the plates we left overnight should have spec-resistance, but they may not have the omega and zif268 unit that we inserted [[#Isothermal Assembly|yesterday]] with isothermal assembly. We performed a PCR on 13 colonies picked from the 50 ul plate to find a colony that contains the w+zif268 insertion. We used the same recipe at our PCR reaction [[#PCR of omega+zif268 with overhangs|on Friday]]. If the insert is present, then we should see a band at 709 bp. We also included 2 positive controls: our isothermal assembly product and our selection strain from which the w+zif268 was originally copied.
| + | |
| - | | + | |
| - | We ran an agarose gel to check the bands:
| + | |
| - | {|
| + | |
| - | |[[File:2011_07_10_transformation_colony_pcr_lane_1-8.png|thumb|Lanes 1-8]]
| + | |
| - | |[[File:2011.07.10_transformation_colony_pcr_lane_9-16.png|thumb|Lanes 9-16]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | None of the colonies displayed any bands, except for primer residues. Lane 16 contained the selection plasmid, on which we performed [[#Gel purification of PCR product|a successful PCR]] on Friday, so we definitely expected a band at 709 bp. Because the positive controls did not have a band at 709 bp, we suspect that something went wrong with the PCR. We plan on re-running the PCR tomorrow.
| + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | <div id="711" style="display:none">
| + | |
| - | ==July 11th==
| + | |
| - | ===Team ZF===
| + | |
| - | We did a miniprep of the 12 colonies we picked Sunday and left overnight in LB. Here are the concentrations of our products:
| + | |
| - | {|
| + | |
| - | !ID
| + | |
| - | !Conc. (ng/ul)
| + | |
| - | !260/280
| + | |
| - | |-
| + | |
| - | |1
| + | |
| - | |6.63
| + | |
| - | |1.27
| + | |
| - | |-
| + | |
| - | |2
| + | |
| - | |12.96
| + | |
| - | |1.53
| + | |
| - | |-
| + | |
| - | |3
| + | |
| - | |9.28
| + | |
| - | |1.41
| + | |
| - | |-
| + | |
| - | |4
| + | |
| - | |7.23
| + | |
| - | |1.31
| + | |
| - | |-
| + | |
| - | |5
| + | |
| - | |12.64
| + | |
| - | |1.55
| + | |
| - | |-
| + | |
| - | |6
| + | |
| - | |10.77
| + | |
| - | |1.48
| + | |
| - | |-
| + | |
| - | |7
| + | |
| - | |17.19
| + | |
| - | |1.73
| + | |
| - | |-
| + | |
| - | |8
| + | |
| - | |39.70
| + | |
| - | |1.78
| + | |
| - | |-
| + | |
| - | |9
| + | |
| - | |55.73
| + | |
| - | |1.81
| + | |
| - | |-
| + | |
| - | |10
| + | |
| - | |24.34
| + | |
| - | |1.72
| + | |
| - | |-
| + | |
| - | |11
| + | |
| - | |25.19
| + | |
| - | |1.68
| + | |
| - | |-
| + | |
| - | |12
| + | |
| - | |38.57
| + | |
| - | |1.74
| + | |
| - | |-
| + | |
| - | |13
| + | |
| - | |27.26
| + | |
| - | |1.75
| + | |
| - | |}
| + | |
| - | | + | |
| - | Then we performed a PCR on the 12 miniprep products, the 12 colonies in LB, as well as 3 controls (negative control, isothermal assembly product, selection strain) to find a colony that has the omega+zif26 insertion. We used the same recipe as [[#PCR of omega+zif268 with overhangs|Friday's PCR]].
| + | |
| - | | + | |
| - | | + | |
| - | We ran a 1% 100 ml. agarose gel at 120V to check the success of the PCR.
| + | |
| - | {|
| + | |
| - | |[[File:7.11.11.omegazif_transformed_annotated.png|thumb|Gel of our colonies, miniprep-ed colonies, and controls to check for w+zif268 insertion]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | The bands were rather thick, so it was difficult to estimate the exact band size. However, the bands appeared a bit below 650 bp, although we expected a size of 709 bp, so we ran an e-gel on some of the colonies to ensure that that bands were indeed the right size.
| + | |
| - | | + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:7.11.11.omegazif_transformed_egel_annotated.png|thumb|Gel to ensure our bands were the correct size (709 bp)]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | The bands were closer to the size that we expected, so we decided that we successfully inserted w+zif268 into the plasmid. We decided to pick colony 9 (which had the highest concentration post-miniprep) and work with that from now on. We made spec plates with 50 ul of colony 9.
| + | |
| - | | + | |
| - | ===Team Wolfe===
| + | |
| - | '''Selection strain transformed with pSR01:'''
| + | |
| - | *both plates had lots of colonies--almost too many. We need to make sure the plasmid is, indeed, in these cells.
| + | |
| - | *Colony PCR:
| + | |
| - | **picked 6 colonies and diluted in 7µL H2O: 3 for PCR template and 4 to be grown up in LB+amp
| + | |
| - | **KAPA protocol using M13_F and M13_R primers (anneal to the vector), 50˚C annealing, 2min elongation
| + | |
| - | **E gel results: lots of bands but rather messy and not exactly what we'd expect. We'll try to verify the presence of the plasmid in a few other ways.
| + | |
| - | [[File: pSR01 test1(labeled).png|thumb|none|pSR01 colony PCR 7/11/11]]
| + | |
| - | *Sequencing:
| + | |
| - | **We sent out the PCR samples with M13_R to Genewiz
| + | |
| - | *Culture PCR:
| + | |
| - | **We used the cultures made with the colonies used for PCR above as template to see if this gives us cleaner results.
| + | |
| - | **same protocol as above except with 1µL of 1:20 dilution of culture as template.
| + | |
| - | *Assuming that the plasmid is indeed in the colonies, we will grow an overnight culture in LB+amp as well as the Wolfe selection strain and tomorrow we will use a growth reader to check their phenotypes in NM
| + | |
| - | '''MAGE round 3 colonies:'''
| + | |
| - | *In case pSR01 does not rescue the selection strain's growth phenotype, we will PCR 96 colonies from the MAGE3 plates and send them out for sequencing tomorrow.
| + | |
| - | **Used 9µL KAPA, His_F and His_R primers, 3µL suspended colony (out of 7--the remaining 4 grown in LB+amp)
| + | |
| - | [[File: 2011.07.11.MAGE 3 96 A1-H1.png|thumb|none|MAGE3 colonies for sequencing 7/11/11]]
| + | |
| - | '''Thio kanZFB:'''
| + | |
| - | *We retried the PCR to put modified nucleotides on the ends of the kan-ZFB-wp-hisura insert using the same procedure as 7/8/11
| + | |
| - | **E gel: reactions didn't work
| + | |
| - | [[File: 2011.07.11.thiokanZFB(labeled).png|thumb|none|thio kan-ZFB PCR 7/11/11]]
| + | |
| - | *Tried again with a longer elongation time (3 min) and let run overnight.
| + | |
| - | | + | |
| - | ===Team TolC===
| + | |
| - | ====Sequencing of rpoZ delete====
| + | |
| - | *Since the sequencing was sent after 3pm on Friday, it wasn't picked up before today (Monday), so we are going ahead with lambda red recombineering of the KAN-ZFB-wp into the ECNR2 genome
| + | |
| - | *glycerol stocks of ECNR2 and ECNR2ΔTolC were made.
| + | |
| - | | + | |
| - | ===Team Web===
| + | |
| - | *Design [https://spreadsheets.google.com/spreadsheet/ccc?key=0ApTl36bX3P7qdFo3SEFXMkNCZ3ZBX1hBQUZqWUpiV2c&pli=1#gid=0 Google Doc]
| + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | <div id="712" style="display:none">
| + | |
| - | ==July 12th==
| + | |
| - | ===Team ZF===
| + | |
| - | ====PCRs of finger ultramers====
| + | |
| - | We received the ultramers for fingers two and three of our 6 experimental expression plasmids. We performed an overlap PCR in order to join the two ultramer pieces and make a whole unit. Ultimately this unit with be assembled with the omega subunit and the spec backbone to make our final expression plasmids. We used the following recipe:
| + | |
| - | *10 ul KAPA
| + | |
| - | *13.5 ul water
| + | |
| - | *0.75 ul forward ultramer
| + | |
| - | *0.75 ul reverse ultramer
| + | |
| - | We had an annealing temperature of 58* and an extension time of 15 seconds.
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.12_nine_ultramers_product_annotated.png|thumb|Gel of our product from the ultramer PCR. Note that the first lane's PCR was done in a single tube, while all the others were done in a strip.]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | However, in addition to the expected band at approximately 300 bp, the bands for our ultramers was extremely streaky, indicating that there may be other contaminants. We decided to perform a PCR purification of these samples, using [[Protocols#PCR purification|our protocols]], and see if there was an improvement in the gel.
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.12_nine_ultramers_product_try2_annotated.png|thumb|Gel of our product after PCR purification ]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | Even after the gel purification, our bands were streaky. We decided to redo the PCR, placing all the reactions in single tubes, performing a touchdown PCR instead. Our annealing temperature started at 70*, repeated for 2 cycles, and then decreased 2*, every two cycles, until it reached 58* (the optimal annealing temperature), where it remained until the 25 cycles were complete. We left this PCR going overnight. (See [[#Results of ultramer touchdown PCR|July 13th]] for the gel image.)
| + | |
| - | | + | |
| - | ====PCR of expression plasmid cross-junction====
| + | |
| - | | + | |
| - | We also performed a second PCR on colony nine (the colony we decided with which we decided to proceed) to confirm that our omega+zif268 in the spec backbone was successfully inserted into the ''E. coli''. The expected product was the cross-junction on the plasmid, approximately 1.4 kb. We used the same recipe as that used on [[#PCR of omega+zif268 with overhangs|July 9th]], except with a forward primer of PZE23G-3581 F and a reverse primer of PZE23G-2133 R, and had a 55* annealing temperature and an extension time of 45 seconds.
| + | |
| - | | + | |
| - | After running an e-gel, we confirmed that our w+zif268 was inserted into the plasmid.
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.12_omega+linker_and_crossjunction_omgzif-spec_plasmid_annotated.png|thumb|'''Lane 3''': Gel of our PCR to check for the cross-junction in colony 9.]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | ====PCR and gel extraction of omega subunit====
| + | |
| - | We performed a PCR to get out the omega subunit+linker, with an annealing temperature of 56* and an extension time of 15 seconds. The miniprep from colony 8 was the template DNA. We will use the product in assembling our final expression plasmids.
| + | |
| - | | + | |
| - | We determined that the PCR was successful after running the sample on an agarose gel, and performed a gel extraction so that we could use the product in our isothermal assembly.
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.12_omega+linker_and_crossjunction_omgzif-spec_plasmid_annotated.png|thumb|'''Lanes 1 and 2''': Gel of our PCR to get omega subunit.]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | The gel extraction was successful, and we had a final concentration of 120.5 ng/ul.
| + | |
| - | | + | |
| - | ====Overlap PCR of OZ052 and OZ123====
| + | |
| - | We performed an overlap PCR using the ultramer pairs that solely encoded the F1, F2, and F3 fingers of OZ052 and OZ123. The annealing temperature was 62* and the extension time was 15 seconds. The results of the PCR can be found below (the products are only in the last two lanes) and the products were also PCR purified before being put into storage at -20*C.
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.12_OZ052_and_OZ123_ultramers_(last_two_lanes)_annotated.png|thumb|'''Last two lanes''': OZ052 and OZ123 products, respectively]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | ===Team Wolfe===
| + | |
| - | '''MAGE round 3 samples:'''
| + | |
| - | *94 sent out for sequencing (numbers go A1-H1, etc.)
| + | |
| - | '''Thio kan-ZFB results:'''
| + | |
| - | *PCR run out on E Gel: not successful. We do not know what is still causing our PCR to fail.
| + | |
| - | '''pSR01-transformed selection strain:'''
| + | |
| - | *We discovered that hisB is a two-function protein, with the N-terminal region acting as an HPase and the C-terminal region as an IDPG. His3 only complements the IDPG activity, so even if our MAGE worked, it might destroy our selection system. The two aa deleted in hisB are in the C terminal region, so maybe they do inactivate it. We emailed Keith Joung whose lab made the selection strain to see how HisB was deleted.
| + | |
| - | **In case this mutation is correct but lambda red in the selection strain still eludes us, we designed MAGE oligos for use in EcNR2 to either delete the two aa or put 3 stop codons after the HPase region.
| + | |
| - | *culture PCR results: cultures 3-4 had a strong band (and 2 and 5 weak ones) around 850-1000bp. It is interesting how different these results are from yesterday's colony PCR gel.
| + | |
| - | [[File: 2011.07.12.pSR01 with phospho primers(labeled).png|thumb|none|pSR01 culture and thio kan-ZFB PCRs 7/12/11]]
| + | |
| - | *Sequencing results: The sequences sent from Genewiz aligned well with the pSR01 plasmid sequence and covered pretty much all of URA3. This implies that the cells did indeed take up our plasmid.
| + | |
| - | *Growth phenotype:
| + | |
| - | **12 colonies were chosen and split between NM (gluc), NM+IPTG (to induce zif268 expression), NM+his (gluc), NM+his+IPTG, LB+amp, LB+amp+IPTG and grown overnight. (Final concentration IPTG=500µM)
| + | |
| - | | + | |
| - | ===Team TolC===
| + | |
| - | *'''MAGE knockout sequencing'''
| + | |
| - | **Received sequencing from the MAGE knockout of rpoZ within the TolC genome
| + | |
| - | **Observed all 24 sequenced attempts and it appears as none of them have the knockout and stop codons placed into genome
| + | |
| - | ***Possible reason behind failure is that one, we may not have been reading the results correctly or two, the MAGE oligo had all three stop codons right next to each other in the oligo and may have needed to be separated by three nucleotides each in order to maximal annealing
| + | |
| - | *'''Lambda Red Recombination of kan-ZFB-wp insertion into MAGE knockouts'''
| + | |
| - | **Plates from the recombination showed small colonies
| + | |
| - | ***With this we picked 6 colonies from the 4 different plated recombination cultures and grew them to saturation
| + | |
| - | ***Began PCR with TolC_Seq_F and TolC_Seq_R primers in order to determine whether recombination was successful
| + | |
| - | ***Procedure (on gradient PCR machine, name: ABC, folder: igem)
| + | |
| - | ****KAPA mixture
| + | |
| - | ****Then 95 C, 5 min
| + | |
| - | ****98˚ C, 20 sec
| + | |
| - | ****60˚ C, 15 sec
| + | |
| - | ****75˚ C, 45 sec
| + | |
| - | ****Repeated cycle for 30 times
| + | |
| - | ****72˚ C, 5 min
| + | |
| - | ****4˚ C, forever
| + | |
| - | ***Tomorrow we plan to run gel
| + | |
| - | ****Length will be:
| + | |
| - | *****450 bp if insertion failed
| + | |
| - | *****1200 bp if insertion of Kan cassette only
| + | |
| - | *****1650 bp if insertion of Kan-ZFB-wp
| + | |
| - | *'''Insertion Construct for TolC (homology-kan-ZFB-wp-homolgy)'''
| + | |
| - | **Began 90 µL total of PCR for the insertion construct since we ran out
| + | |
| - | *If gels show failed PCR tomorrow we will start from normal ENR2 without any MAGE and begin with lambda red recombination and possibly make new MAGE primer and then knockout rpoZ
| + | |
| - | | + | |
| - | ===Team Web Design===
| + | |
| - | *Met with Harvard biosafety officer, Sidney
| + | |
| - | **Send to him: list of strains, plasmids that we are using
| + | |
| - | **<font color=red>'''''Note to all iGEM-ers:''''' We will all be attending a biosafety seminar on Friday, July 15th from 10am to 12n in Biolabs 1058. </font>
| + | |
| - | | + | |
| - | ::In case you haven't already, please see Alain's message below:
| + | |
| - | | + | |
| - | ::Every member of the Harvard iGEM team (unless you are absent on Friday) has to attend the next lab Safey/Biosafety Blood born pathogens training this coming Friday 7/15/2011 from 10 am to 12 noon. The training takes place in the Biological laboratories (16 Divinity Ave) room 1058. If you cannot go and have a very good reason let me (''Alain'') know. If you attended a similar training session on the Cambridge campus or at the Medical school you don't have to attend. Otherwise you have to go.
| + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | <div id="713" style="display:none">
| + | |
| - | ==July 13th==
| + | |
| - | ===Team ZF===
| + | |
| - | ====Results of ultramer touchdown PCR====
| + | |
| - | | + | |
| - | We ran an e-gel on the product of our touchdown PCR. Once again we have our band at 344 bp. It is still slighly streaky, but much less than our [[#PCRs of finger ultramers|earlier PCR reaction]]. There are some faint bands smaller than our expected product, which are likely the ultramers we used in the reaction. We'll use the product for our isothermal assembly later today.
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.13_ultramers_annotated.png|thumb|Gel of our ultramer touchdown PCR product]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | ====Isothermal assembly and transformation of expression plasmids====
| + | |
| - | We followed [[Protocols#Isothermal assembly|our protocol]] for isothermal assembly, using the following recipe:
| + | |
| - | *0.63 ul backbone (at 157.4 ng/ul to get 100 ng)
| + | |
| - | *0.13 ul omega+linker (at 120.5 ng/ul)
| + | |
| - | *0.50 ul ultramer (~30 ng/ul)
| + | |
| - | *1.24 ul water
| + | |
| - | *7.5 ul isothermal assembly solution
| + | |
| - | | + | |
| - | The sizes of the pieces we put together were the following:
| + | |
| - | *Backbone: 2.2 kb
| + | |
| - | *omega+linker: 352 bp
| + | |
| - | *ultramer: 344 bp
| + | |
| - | | + | |
| - | Note: We also made a negative control of the isothermal assembly, only putting in the spec backbone without omega+linker or ultramer.
| + | |
| - | | + | |
| - | We then performed transformations to insert our isothermal assembly products into ''E. coli'', following our [[Protocols#Cultures|usual protocol]]. However, we ran out of electrocompetent cells, so only samples 78 - 82 were done with electroporation while 77, 83-85, and the negative control were done with chemical transformation, using [[Protocols#Chemical transformation|these protocols]] (the samples are named after the reverse ultramer ID in the primer index that were used to generate the ultramer sequences).
| + | |
| - | | + | |
| - | We then plated 1 ul of the cells on spec plates and left them overnight at 37*. Because we ran out of our own spec plates, we also had to use LB plates that had spec added to it, and spec plates that were made in April.
| + | |
| - | | + | |
| - | ===Team Wolfe===
| + | |
| - | '''∆HisB∆pyrF∆rpoZ+pSR01 growth results:'''
| + | |
| - | *When no IPTG was present, the bacteria grew in LB+amp, NM+his, and NM, showing that the plasmid rescues the selection stain's growth phenotype! This implies that our strain had the right genotype all along. However, when IPTG was added to the media, the strain only grew well in LB. This is very odd, since IPTG was supposed to induce zif268 and thus His3 expression. We will try a couple things to make sure nothing went wrong:
| + | |
| - | **redo cultures in the same pattern as yesterday--perhaps IPTG+/- were mislabeled
| + | |
| - | **inoculate cultures of ∆HisB∆pyrF∆rpoZ +/- pSR01 so that Vatsan can use a plate reader to compare their growths
| + | |
| - | *Joung wrote back and said that the HisB deletion is in fact 6 bases in frame. He also mentioned something about a lac deletion which we are looking into.
| + | |
| - | '''MAGE3 sequencing results:'''
| + | |
| - | *none of the colonies had the stop codons. This may be because mutS is not knocked out, and it can correct strands that have up to 5 bases mismatched.
| + | |
| - | '''Kan-ZFB for lambda red:'''
| + | |
| - | *Noah did lambda red in EcNR2 and the plate was covered in crystalline patterns that may be cells. We will test this by:
| + | |
| - | **Restreaking some of these "colonies" onto a fresh kan plate to get the bacteria more spread out
| + | |
| - | **Streaking a kan plate with pKD46 (from glycerol stock) to see if the kan is effective
| + | |
| - | **Taken a streak off the plate, diluted it in 200µL H20, and used 1µL as template for PCR
| + | |
| - | ***KAPA, 1529620-flanking primers, 90 sec elongation and 56 anneal, 25 cycles
| + | |
| - | ***Results: 300bp band--the streaks on the plate are bacteria, but they do not have the insert.
| + | |
| - | [[File: 2011.07.13 Lambda red streaky plate(labeled).png|thumb|none|1529620 locus PCR of EcNR2 lambda red 7/13/11]]
| + | |
| - | *We also retried the thio kan-ZFB PCR using some new primer aliquots. Once again it failed.
| + | |
| - | [[File: 2011.07.13.Phospho thio 4(labeled).png|thumb|none|Thio kan-ZFB PCR 7/13/11]]
| + | |
| - | *Because this PCR keeps failing, we're going to try a different way and do two separate and shorter PCRs to be joined later in an overlap PCR:
| + | |
| - | **PCR of kan cassette: kan cassette as template (diluted 6/16 aliquot), kan F phosphothio and kan R primers, KAPA, 56˚C anneal and 90 sec elongation
| + | |
| - | **PCR of ZFB-wp-hisura: ZFB-wp-hisura as template (diluted 6/16 aliquot), ZFB-wp-f and URA3 R phosphothio primers, KAPA, 60˚C anneal and 90 sec elongation
| + | |
| - | | + | |
| - | ===Team TolC===
| + | |
| - | *'''PCR ECNR2'''
| + | |
| - | **Ran PCR on four colonies from lambda red recombination results, ECNR2 liquid culture from Church lab, and colony from original ECNR2 plate
| + | |
| - | ***Grew each up in LB Amp liquid culture
| + | |
| - | ***Tomorrow the gel will tell us whether the insertion was successful and also if the ECNR2 cultures are pure
| + | |
| - | *'''PCR Noah's MAGE on rpoZ knockout'''
| + | |
| - | **Picked 15 colonies from each plate with ECNR2 without TolC and ECNR2 with TolC
| + | |
| - | ***Created liquid cultures of all 30 colonies picked and growing up over night
| + | |
| - | **PCR will be ready to send out tomorrow for sequencing
| + | |
| - | *'''More kan-ZFB-wp insertion construct needed'''
| + | |
| - | **Ran PCR in order to obtain more kan-ZFB-wp insertion construct from the kan-ZFB-wp-His3-PyrF construct
| + | |
| - | **Placed on gel and used gel extraction in order to purify
| + | |
| - | ***10.7 ng/
| + | |
| - | | + | |
| - | [[File: 2011.07.14.TolCinsertionconstruct(labeled).png|thumb|none|KAN-ZFB-wp construct with tolC homology 7/13/11]]
| + | |
| - | | + | |
| - | ===Team Web Design===
| + | |
| - | *Safety, safety, safety!
| + | |
| - | *Received safety confirmation letter
| + | |
| - | *Finalized project description
| + | |
| - | *Updated our iGEM wiki page for the upcoming deadlines (Project description and safety, due July 15th)
| + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | <div id="714" style="display:none">
| + | |
| - | ==July 14th==
| + | |
| - | ===Team ZF===
| + | |
| - | ====Results of the expression plasmid transformation====
| + | |
| - | Only samples 78 to 82 had colonies after overnight incubation--these were the same colonies that were electroporated, instead of chemically transformed. Because we only added 1 ul of our cells to the plates, it is possible that this was not enough sample for us to see colonies, especially since chemical transformation is not as efficient as electroporation. The samples on the LB+spec plates had what appeared to be unspecific growth around the edges. This may be because the spec did not diffuse evenly. We will be re-plating all of our samples later today, on new LB+spec plates.
| + | |
| - | | + | |
| - | ====PCR of product from isothermal assembly of expression plasmid====
| + | |
| - | We also ran a PCR on the product of the three-part isothermal assembly (omega+linker, ultramers, spec backbone). We used the cross-junction primers and the same recipe as our PCR [[#PCR of expression plasmid cross-junction|on Tuesday]]. Once again we expected a product of 1.4 kb.
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.14_9_ultramer_isotherm_asm_rxn_with_junction_primers_annotated.png|thumb|1.2% E-gel of our ultramer isothermal assembly reaction product]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | There were fairly sharp bands underneath the band we expected. Additionally, the expected 1.4 kb band was not visible for samples 80 and 81, although they were successfully grown on plates. Sample 85 had no bands, not even primers.
| + | |
| - | | + | |
| - | Since 80 and 81 grew on our plates yesterday, we continued with our plan of re-plating our original transformations, especially since the chemically transformed cells do have the appropriate isothermal assembly product to insert. We re-plated all the transformations on the LB+spec plates made today, and will leave them overnight in the 37* incubator. We also made cultures from the colonies on plates 78-82 to mini-prep tomorrow.
| + | |
| - | | + | |
| - | ====Rerunning of PCR products (ultramers and isothermal assembly)====
| + | |
| - | In addition, we decided to see if the streaking was possibly caused by the gel instead of a PCR problem, and so we ran three of our products in a 1% agarose gel instead of an E-gel, with the results below.
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.14_ultramers.pcr1_touchdown_pcr_iso_assm_annotated.png|thumb|Products ran in an agarose gel. Note that wells 78-80 on the left side of the gel are empty due to sample mysteriously floating out of the well.]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | ===Team Web Design===
| + | |
| - | *Intro video: brief (~1:30) Flash video explaining our project (Our project in 30 sec? 1 min?)
| + | |
| - | *Interview Joung/Persikov-- videotape (documentary style) for Human Practices page
| + | |
| - | *Human Practices
| + | |
| - | **Broader societal impacts
| + | |
| - | **Gene therapy, personalized medicine
| + | |
| - | ===Team Wolfe===
| + | |
| - | '''thio kan and thio ZFB-wp-hisura PCR:'''
| + | |
| - | *Ran overnight PCR of kan cassette and ZFB-wp-hisura on E gel: looks great!
| + | |
| - | [[File: 2011.07.14.thiokan+thioZFBhisura(labeled).png|thumb|none|thio kan and thio ZFB-wp-hisura PCR 7/14/11]]
| + | |
| - | *PCR purification with Qiagen kit:
| + | |
| - | **added an additional 300µL buffer PB to adjust the pH
| + | |
| - | **thio kan cassette: 59.9ng/µL 260/280=1.90
| + | |
| - | **thio ZFB-wp-hisura: 100.9ng/µL 260/280=1.89
| + | |
| - | *Overlap PCR:
| + | |
| - | **4 rxns: two used undiluted template (60ng thio kan cassette and 101ng thio ZFB-wp-hisura), two used template diluted 1:10
| + | |
| - | **KAPA, 56˚C anneal, 90 sec elongation, 25 cycles
| + | |
| - | **primers (kan f phosphothio and ura3 r phosphothio) added after 10 cycles
| + | |
| - | **Results: PCR worked but with tons of side products so we will try to optimize the procedure. Expected product = 3Kb, notable side products = 2Kb, 4Kb. N.B. The samples were accidentally loaded while the gel was running so they are staggered on the gel.
| + | |
| - | [[File: 2011.07.14.thio kanZFBhisura overlap(labeled).png|thumb|none|thio kan-ZFB-wp-hisura overlap PCR (with loading error)]]
| + | |
| - | *Gel stab PCR
| + | |
| - | ** DNA from the 3Kb e-gel band were removed using the stabbing technique and placed in 4 respective reaction tubes (diluted in 20µL water)
| + | |
| - | ** The 4 samples were separated into template dilutions of 20X, 100X, 200X, and 400X and phosphothio primers for PCR to amplify the desired 3Kb kan-ZFB-wp-hisura sequence. PCR results will be reported tomorrow.
| + | |
| - | *Template PCR followed by PCR with primers:
| + | |
| - | # PCR just the template (1µL diluted thio kan cassette and 1µL diluted thio ZFB-wp-hisura) with no primers, KAPA, 12 cycles, 60˚C anneal, 90 sec elongation
| + | |
| - | # take PCR product and use it as template for new PCR either undiluted, diluted 1:4, 1:16, 1:64, 1:256; kan f phosphothio and ura3 r phosphothio primers, 25 cycles, 59˚C anneal, 90 sec elongation
| + | |
| - | '''pSR01 phenotype:'''
| + | |
| - | *Just to be sure, we will assemble cultures in a flat-bottomed 96 well plate and use a plate reader to track the growth curves.
| + | |
| - | *2 colonies ∆HisB∆pyrF∆rpoZ, 10 colonies transformed with pSR01
| + | |
| - | *media: LB+amp, NM+his, NM, NM+3-AT (1mM), LB+amp+IPTG(100µM), NM+his+IPTG, NM+IPTG, NM+3-AT+IPTG
| + | |
| - | *plate reader: absorbency of 600nm, measurements every 10 min, shaking 560 sec before each measurement (settings saved in iGEM folder on computer in tissue culture room)
| + | |
| - | | + | |
| - | ===Team TolC===
| + | |
| - | *PCR done of 4 colonies picked from KAN plate (with ECNR2 after recombineering with the KAN-ZFB-wp construct. Running an E-gel showed that the insert was not present. Since the KAN plates were old, it might be speculated that the plates were old, and so, the colonies were not selected for KAN resistance.
| + | |
| - | **Primers were TolC_seq_F and TolC_seq_R.
| + | |
| - | ***Without insertion these two primers will create 449 bp sequence, and with insertion length will be 1650 bp
| + | |
| - | [[File: 2011.07.06.TolCinsertionconstruct+negativecontrol(labeled).png|thumb|none|TolC insert check 1 7/14/11]]
| + | |
| - | **Some of the original culture after recombineering, 1ml solution spun down, and resuspended in 100μl of LB, and plated on new KAN plates. To check, one of the old KAN plates was plated the same way and put in at 2:30pm. (These plates will be checked tomorrow morning.)
| + | |
| - | *PCR done of a few of the rpoZ MAGE with stop codon insertions. Oligo name: rpoZ_MAGE_KO (Noah's oligo) (Noah's plates, after two cycles of MAGE, on both ECNR2 and ECNR2ΔTolC). The side product in the gel could be due to a low annealing temperature of 53°C, so we should do a gradient PCR (e.g.55-66°C) to get the most product the next time.
| + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | <div id="715" style="display:none">
| + | |
| - | ==July 15th==
| + | |
| - | ===Team Wolfe===
| + | |
| - | '''Thio kan-ZFB-wp-hisura optimizing PCR results:'''
| + | |
| - | *The gel stab PCR produced a product of the right size, but the 2kb side product was still brighter. The 2-PCR protocol also still had substantial side products but at least they did not completely overshadow the real product. We will accordingly go ahead and do a gel extraction on the 2-PCR samples so that we can try lambda red with them next week.
| + | |
| - | [[File: 2011.07.15.thiokan PCR optimization(labeled).png||thumb|none|Different attempts and thio kan-ZFB-hisura PCR 7/15/11]]
| + | |
| - | '''pSR01 growth phenotype results:'''
| + | |
| - | *Unfortunately the computer hooked up to the plate reader spontaneously restarted during the night to download updates, so we lost all the data. Just by looking at the plate, it still seems like IPTG is inhibiting growth. The ∆HisB∆pyrF∆rpoZ controls also did not grow, probably because the plate the colonies were taken from was too old, so we will restreak from the glycerol stock and set up the plate reader again this weekend.
| + | |
| - | | + | |
| - | ===Team ZF===
| + | |
| - | ====PCR of electro comp expression plasmids====
| + | |
| - | We performed a mini-prep of the cultures that we grew overnight of the electrocompetent cells. We then performed a PCR on the product of the mini-prep, checking for the 1.4 kb cross junction to confirm the presence of our omega+ultramers+spec plasmid. We used the same recipe as the PCR we performed [[#PCR of expression plasmid cross-junction|on Tuesday]].
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.15_electro_miniprep_and_cultures_annotated.png|thumb|Description of gel below.]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | No bands appeared for PCR peformed on a 1 to 10 dilution of the cultures. Miniprep product from colonies 78, 79, and 82 had the expected band at 1.4 kb, while 80 and 81 had a band that was slightly smaller than the the expected product size. We saw a similar band in our [[#PCR of product from isothermal assembly of expression plasmid|PCR of the isothermal assembly reaction product]]. We are unsure exactly what is causing this band. If either the omega subunit or ultramers were missing from the plasmid, a similar sized product would result since both are ~350 bp. We are curious if a plasmid can still be formed if one of these components were missing.
| + | |
| - | | + | |
| - | We checked the concentrations of our miniprep product to see if that would explain our problem.
| + | |
| - | ID Conc. (ng/ul) 260/280 1.4 kb band?
| + | |
| - | 78 16.12 2.10 Y
| + | |
| - | 79 41.64 1.64 Y
| + | |
| - | 80 5.84 2.01 N
| + | |
| - | 81 61.24 1.62 N
| + | |
| - | 82 20.25 1.72 Y
| + | |
| - | | + | |
| - | We can't tell whether the concentration of the miniprep product contributed to the strange band we saw. 81 had the highest concentration but still didn't work. We decided to try plating 4 more colonies from 80 and 81, to see if any of them have the appropriate band.
| + | |
| - | | + | |
| - | We also decided to make final plates of cultures 78, 79, and 82, using 2 ul of culture and 50 ul of LB+spec. They were plated on new spec plates. We will incubate them at 37* overnight.
| + | |
| - | | + | |
| - | ====Checking the expression plasmid plates====
| + | |
| - | All the plates had colonies on them, although there were still areas of unspecific growth, presumably because of uneven dilution of the spec into the LB plates.
| + | |
| - | | + | |
| - | We made cultures of all the chem comp cells (77, 83, 84, 85) and the electro comp cell that did not have the correct insertion (80, 81). We picked 4 colonies from each plate, resulting in a total of 24 cultures. We have left them in the 37* shaker overnight, and tomorrow plan on performing a miniprep and PCR to check for the cross-junction.
| + | |
| - | | + | |
| - | ====PCR to check for omega+ultramers====
| + | |
| - | We performed a PCR on the five electro comp miniprep products, to find out what may be missing. We used the omega_F+pLacO (index: 73) as our forward primer, and each of the respective reverse ultramers as our reverse primers (index: 78-82). We had an annealing temperature of 58* and an extension time of 30 seconds, and the expected product size is 646 bp.
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.15_Team_TolC(lanes_2-7)_and_omega-ultramer_plasmid_pcr_annotated.png|thumb|Team ZF results in lanes 8-12]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | Strangely, we did not get results for sample 78 (FH bottom), even though we got a band there in a PCR earlier today that used the junction primers. We got positive results for samples 79 and 82, and everything else seems to check out with these two samples (bands present in the correct size with junction primers from both isothermal assembly and miniprep templates). However, we are still having problems with samples 80 and 81, which may be a problem with the colonies that we picked. For now, we will assume that the isothermal assembly worked, due to colonies growing on the spec plates for samples 80 and 81, and that we had simply picked bad colonies. We will find out tomorrow if other colonies from the same plate give us the same results.
| + | |
| - | </div>
| + | |
| - | | + | |
| - | <div id="716" style="display:none">
| + | |
| - | ==July 16th==
| + | |
| - | ===Team ZF===
| + | |
| - | ====Colony minipreps====
| + | |
| - | Minipreps were performed on all of the samples except for a few where nothing grew in the LB/spec media. These exceptions included 85.3, 85.4, 84.2, 81.2, and 81.3. Possible reasons for no growth for these samples may include: (1) The colony was somehow not a transformed colony and was a cheater (possibly picked from a spot where there was a smaller amount of spec in the unevenly distributed dish), and (2) the colony did not get picked up properly with the pipet tip.
| + | |
| - | | + | |
| - | The concentrations of the DNA after miniprep were somewhat low, and a full table of these concentrations and DNA purities can be found below:
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.16_Colony_Miniprep_Conc.jpg|thumb|Plasmid concentrations]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | ====PCR on minipreps to check cross junction====
| + | |
| - | PCR was done on the miniprep products to check for the cross junction length, expected band size of ~1.4 kb. The protocol used was the same as [[#PCR of expression plasmid cross-junction|the previous time]]. In addition to PCR-ing the miniprep products, a positive control was also included, which was the finished Zif268 + omega + spec backbone plasmid (this was the first successful junction PCR we obtained, and so it should work again).
| + | |
| - | | + | |
| - | ====Replating final samples for 78, 79, 82====
| + | |
| - | Strangely, nothing grew on the plates that we seeded yesterday with 2ul of bacteria. We seeded plates again for samples 78, 79, and 82 with 15ul of transformed bacteria instead, and hopefully these should grow by tomorrow.
| + | |
| - | </div>
| + | |
| - | | + | |
| - | <div id="717" style="display:none">
| + | |
| - | ==July 17th==
| + | |
| - | ===Team ZF===
| + | |
| - | ====Replating final samples, take 2====
| + | |
| - | We realized that the plates we seeded yesterday (July 16) were seeded from the transformation bacteria tubes, which has a mix of bacteria that may/may not have taken up the plasmid. These plates therefore cannot serve as the final stock plates. We took the E. coli from our initial colony picking and put them in LB + spec to grow to saturation tomorrow, where we will replate again with those bacteria (for which the good PCR results should correlate).
| + | |
| - | ====Gel analysis of miniprep junction PCR====
| + | |
| - | The miniprep junction PCR products from yesterday were ran on an agarose gel, with the results below:
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.17_miniprep_junction_pcr_darker_annotated.png|thumb|PCR Results, expected band at ~1.4 kb]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | While all the bands are between 1000 and 1650 kb, there are slight variations in the size for which we are unable to account. We will send these out for sequencing, to try to figure out what is going on.
| + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | <div id="718" style="display:none">
| + | |
| - | ==July 18th==
| + | |
| - | ===Team ZF===
| + | |
| - | ====Sending expression plasmids out for sequencing====
| + | |
| - | We decided to send out the expression plasmids that had bands at approximately the correct size out for sequencing. We originally wanted to sequence the plasmids, but our miniprep concentrations were not high enough. Apparently, we should be getting high yields from minipreps, but thus far our yields have been around 50 ng/ul at the highest.
| + | |
| - | | + | |
| - | We decided to sequence the unpurified PCR products instead. In order to do this we had to re-do the PCRs for Zif268, FH bottom (78), CB top (79), Myc 981 (82). Below are our 1% e-gel results, verifying that we had appropriately sized bands:
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.18_junction_primer_pcr_for_seq_annotated.png|thumb|Gel of cross junction of Zif268, FH bottom, CB top, Myc 981]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | Our final samples sent out for sequencing are:
| + | |
| - | Index Sample ID Index Sample ID
| + | |
| - | 1 77_1 11 80_3
| + | |
| - | 2 77_2 12 80_4
| + | |
| - | 3 77_3 13 -
| + | |
| - | 4 77_4 14 83_2
| + | |
| - | 5 84_1 15 83_3
| + | |
| - | 6 84_4 16 83_4
| + | |
| - | 7 81_1 17 Zif268
| + | |
| - | 8 81_4 18 FH_bottom
| + | |
| - | 9 80_1 19 CB_top
| + | |
| - | 10 80_2 20 Myc_981
| + | |
| - | | + | |
| - | ====Making more cultures and plates of our expression plasmids====
| + | |
| - | We picked two more colonies from plate 85 today and made 5 ml cultures from them. We also made 2 fresh cultures from the old 85 cultures, by taking 50 ul of old culture and 5 ml LB+spec. We will leave them overnight in the 37* shaker and perform minipreps and PCRs on them tomorrow.
| + | |
| - | | + | |
| - | We made positive control cultures from 81.1, 81.4, 80.1, and 80.2, in order to test our minipreps tomorrow. They reactions have a wide range of concentrations post-miniprep, and we are curious to see if the same will happen after performing a miniprep a second time.
| + | |
| - | | + | |
| - | Another way to figure out if our expression plasmids contain the appropriate insert is by using a restriction enzyme to cut it and then performing a PCR on the linear DNA. We had previously inserted 2 cutsites on our plasmids, Bsa1 (for insertion of finger 1) and Xba1. The lab already has Xba1, and we ordered Bsa1. Xba1 can be affected by methylation if it is preceded by 'GA' or followed by 'TC' [http://www.neb.com/nebecomm/products/faqproductR0145.asp#874]; we checked and our sequences should not be affected.
| + | |
| - | | + | |
| - | | + | |
| - | We also re-plated 78, 79, and 82 today, one set of plates with 1 ul of culture, and the other set with 10 ul.
| + | |
| - | | + | |
| - | ====Creating the 96 well test plate====
| + | |
| - | | + | |
| - | ===Team Wolfe===
| + | |
| - | '''SNOWMAN!'''
| + | |
| - | [[File: snowman1.jpeg|left|Our snowman Persikov]]
| + | |
| - | [[File: snowman3.jpeg|none|Brandon with his creation]]
| + | |
| - | | + | |
| - | '''EcNR2 lambda red (Noah's prep):'''
| + | |
| - | *Noah remade the kan-ZFB-wp-hisura insert for us with the kan cassette in the opposite orientation from the his3 and ura3 genes. (The kan cassette is under a strong promoter and does not have a transcriptional terminator, so it's possible that RNA polymerase might keep transcribing through the end of kan and into his3.) He also used it for lambda red in EcNR2, and a couple of plates look like they may have a couple colonies.
| + | |
| - | *picked 4 colonies, diluted in 20µL H20 and used 1µL as template for 1529620 locus PCR. The rest was grown in LB+kan.
| + | |
| - | '''Lambda red with Noah's kanZFBhisura:'''
| + | |
| - | *We recently discovered that the kan plates we had been using were very old, and that might be why we got that weird crystalline lawn of colony-like things. We will retry lambda red in the ∆hisB∆pyrF∆rpoZ +pKD46 strain and see if this solves the problem
| + | |
| - | *1.5mL cells divided in two: half for kan-his-ura (300ng) and half as a negative control (water)
| + | |
| - | '''Thio insert:'''
| + | |
| - | *In case lambda red also does not work, we will try to add phosphothioate bases to the insert, but only to the lagging strand 5' end (so only the Ura3 side). Since our primers will have very different melting temps, we will try 2 different reactions:
| + | |
| - | *Touchdown PCR:
| + | |
| - | **template: Noah's kan-ZFB-hisura (163ng/µL) diluted to ~10ng per reaction (4 reactions)
| + | |
| - | **primers: kan_f+homology, Ura3 R phosphothioate
| + | |
| - | **2 cycles at each annealing temperature (67, 66...57) for 11 cycles followed by 15 more cycles at 57; 3 min elongation
| + | |
| - | *Gradient PCR:
| + | |
| - | **same as above but with 12 reactions, annealing temperatures from 57-67˚C
| + | |
| - | *Results: most of the gradient reactions produced a faint but clean band at 3kb. The touch down PCR had a stronger 3kb band but also lots of side products.
| + | |
| - | [[File: 2011.07.18.thiokan grad1(labeled).png|thumb|left|Thio kan-ZFB-hisura (Noah's) gradient part 1 7/18/11]]
| + | |
| - | [[File: 2011.07.18.thiokan grad2+td(labeled).png|none|Thio kan-ZFB-hisura (Noah's) gradient part 2 and touchdown 7/18/11]]
| + | |
| - | | + | |
| - | ===Team TolC===
| + | |
| - | ====Primers====
| + | |
| - | *Created and ordered primers in order to test the presence of MAGE rpoZ knockout oligo
| + | |
| - | **Primers made are named
| + | |
| - | ***rpoZ_MAGE_KO plus 4, rpoZ_MAGE_KO minus 4, which have all 4 of the stop codons
| + | |
| - | ***rpoZ_MAGE_KO plus 2, and rpoZ_MAGE_KO minus 2, which have only the first two stop codons.
| + | |
| - | | + | |
| - | ====Kan Plates====
| + | |
| - | *Performed control experiment on Kan plates by growing up liquid culture from glycerol stock and then spreading on Kan plate in order to determine the speed of growth and intensity of the presence of Kan on the plates
| + | |
| - | *Will check on these tomorrow.
| + | |
| - | | + | |
| - | ====Kan-ZFB-wp insertion====
| + | |
| - | *Picked 5 colonies from ZF090 and ZF091 plates for a total of 10, ZF092 had too few colonies to pick from
| + | |
| - | **Placed colonies in 20 μL of water and used 1 μl for a 20 μL PCR reaction
| + | |
| - | **Then took 10 μL from 20 μL dilution and added to 100 μL of LB in 96 well plate and put in 30°C fridge, at 3:19 pm
| + | |
| - | **Same protocol as [[Protocols#PCR|KAPA PCR]] with the following changes made
| + | |
| - | ***Step 3, gradient for annealing, 55°C - 77°C, for a range of 55,57,59,61 and 63°C for the two rows of 5 PCR tubes each (placed leftmost)
| + | |
| - | ***Step 4, Extension, 1 min
| + | |
| - | ***Step 5, 25 cycles total
| + | |
| - | **Name: TOLCGRAD, PCR5 machine.
| + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | <div id="719" style="display:none">
| + | |
| - | ==July 19th==
| + | |
| - | ===Team ZF===
| + | |
| - | ====Checking on plates 78, 79, 82====
| + | |
| - | Both the 10 ul and 1 ul essentially had a lawn of bacteria growing on them. We re-plated these samples, using 1/100 ul and 1/10000 ul of bacteria in 50 ul of LB. We will leave these plates to incubate overnight at 37*.
| + | |
| - | | + | |
| - | We also made a culture from the old 85 culture, which we will plate tomorrow.
| + | |
| - | | + | |
| - | ====Minipreps of colonies and controls====
| + | |
| - | Our minipreps from [[#Colony minipreps|Saturday]] had low yields, especially for culture 85. We suspect this is why the PCR [[#Gel analysis of miniprep junction PCR|on Sunday]] did not work for this sample. Unfortunately, 85.3 and 85.4 did not grow even after being left in the shaker overnight. These were two cultures we made by picking new colonies from plate 85. This may have occurred because we did not actually pick the colony, or the colony was a cheater. Thus, we performed minipreps on 80.1, 80.2, 81.1, 81.4 (the controls, chosen for their wide range of concentrations after our first miniprep), and 85.1 and 85.2.
| + | |
| - | | + | |
| - | In order to figure our why our minipreps were resulting in unexpectedly low yields and increase our yields:
| + | |
| - | *We tested the pH of our water and Buffer EB.
| + | |
| - | **The pH of our aliquot of water was ~4-5 (too acidic...we are unsure why)
| + | |
| - | **The pH of our Buffer EB was ~7.
| + | |
| - | ***Ideally, the DNA should be eluted by a solution of pH 7, so we decided to use Buffer EB during the elution step.
| + | |
| - | *We also checked the copy number of the origin of replication used in our plasmids. ColE1 apparently has a low copy number, which might also explain our low yields.
| + | |
| - | *We had made 5 ml of culture in an attempt to increase our yields.
| + | |
| - | | + | |
| - | Our yields were larger than our first round of minipreps, although the purities were rather low. We decided to proceed anyway.
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File: 2011.7.18_Miniprep_redo.jpg |thumb|Concentration and purity of our miniprep product]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | ====PCR of miniprep products====
| + | |
| - | We then ran a PCR for the cross-junction on all 6 (80.1, 80.2, 81.1, 81.4, 85.1, 85.2) samples, using [[#PCR of expression plasmid cross-junction|our usual cross junction protocol]].
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.19_miniprep_test_annotated.png|thumb|Gel results, expected product size of 1.4 kb]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | We saw bands for samples 80.1, 81.1, 81.4, 85.1, and a very faint band for 85.2. The strange thing about these bands is that they appeared to be right around 1650 bp, larger than our 1.4 kb target size. We are unsure of the identity of these bands and will send them out for sequencing.
| + | |
| - | | + | |
| - | ====Starting a new plate for sample 85====
| + | |
| - | Since we have been having such bad luck with sample 85, we seeded 2 ml of LB+spec with 100 ul of the transformed bacteria for sample 85. This is in preparation for making a new plate of sample 85 (the old plate was not a good spec plate and had lots of unspecific growth as well as very few colonies, so if we used this plate for more colony picking our choices would be very limited). This new plate should also give us more luck with picking colonies and having them actually grow in the LB+spec.
| + | |
| - | | + | |
| - | ====Designing ultramers for Zif268, OZ052, and OZ123====
| + | |
| - | In preparation for designing our 96-well practice plate, we designed ultramers for our three positive controls Zif268, OZ052, and OZ123. These ultramers contain the Type II binding sites as well as the F2/F3 fingers. Since the [[Lab_Notebook#Design_of_Plate_Practice_Sequences|practice plate]] will contain oligos that encode for F1, we can use these three positive controls to practice and ensure that we secure the technique of using the Type II binding sites to swap in F1 into our expression plasmids. The ultramers have not yet been ordered.
| + | |
| - | | + | |
| - | ===Team TolC===
| + | |
| - | ====PCR to test insertion of KAN-ZFB-wp into ECNR2====
| + | |
| - | *Made 26 dilutions of culture from three different recombineering plates
| + | |
| - | **10 cultures from Kan_tolC_locus_F1 (ZF090 plate) primer recombination
| + | |
| - | **10 cultures from Kan_tolC_locus_F2 (ZF091 plate) primer recombination
| + | |
| - | **5 cultures from Kan_tolC_locus_F3 (ZF092 plate) primer reecombination
| + | |
| - | **1 culture from an old Kan plate with recombination from last week
| + | |
| - | *5 of ZF090, 6 of ZF091, and 5 ZF092 were placed in a PCR with 58˚C annealing temperature
| + | |
| - | *5 of ZF090, 4 of ZF091, and 1 from old plate were run in a PCR with 62˚C annealing temperature
| + | |
| - | *primers used were TolC_seq_F and TolC_seq_R
| + | |
| - | [[File: 2011.07.19.Noahkan-ZFBinsertionsuccessPCR.png|thumb|none|EcNR2 lambda red 1529620 PCR 7/19/11]]
| + | |
| - | | + | |
| - | [[File: 2011.07.19.Noahkan-ZFBinsertionsuccessPCR2.png|thumb|none|EcNR2 lambda red 1529620 PCR 7/19/11]]
| + | |
| - | | + | |
| - | *Chose colonies based on the intensity of the bands and grew up cultures of ZF091-2d, ZF091-4d, ZF090-1d, ZF091-5d, ZF090-1s, ZF090-2s, ZF090-3s, and ZF091-3s
| + | |
| - | | + | |
| - | ====Sequencing of Kan-ZFB-wp insertion====
| + | |
| - | *Prepared 16 PCR products of the 8 cultures we chose to grow up in order to send out for sequencing
| + | |
| - | **Used TolC_Seq_F and ZFB-wp-F primers
| + | |
| - | **Bands looked a little small when ran out gel for longer
| + | |
| - | | + | |
| - | ====New MAGE oligo====
| + | |
| - | *Constructed a new oligo for MAGE knockout of rpoZ, with oligos more than 4 mutations and substitutions the efficiency drops from 15% down to below 5%, so we created a rpoZ knockout oligo with only 2 substitutions
| + | |
| - | **The new MAGE rpoZ knockout is rpoZ_MAGE_Subs
| + | |
| - | *Also ordered primers to test the presence of the knockout rpoZ_MAGE_Subs1+, rpoZ_MAGE_Subs1-, rpoZ_MAGE_Subs2+, rpoZ_MAGE_Subs2-
| + | |
| - | | + | |
| - | ===Team Wolfe===
| + | |
| - | '''EcNR2 lambda red colony PCR results:'''
| + | |
| - | *PCR showed that colonies 1-3 had the insert while 4 did not. Yay! (Note that in the gel image, we accidentally cut off the 300bp band in lane 4)
| + | |
| - | *We will grow up a reinoculated culture of colony 1 for a glycerol stock. If the Wolfe strain keeps not working, we at least have the insert in an EcNR2 line and can hopefully MAGE out the other genes without too much difficulty.
| + | |
| - | [[File: 2011.07.19.EcNR2 lambda red insert(labeled).png|thumb|none|EcNR2 lambda red 1529620 PCR 7/19/11]]
| + | |
| - | '''pSR01 growth phenotype:'''
| + | |
| - | *We will try to used the plate reader again to compare the growth phenotypes of ∆HisB∆pyrF∆rpoZ and ∆HisB∆pyrF∆rpoZ+pSR01. Used the same media as last time but since we're using a 48 well plate, each well has 250µL media. We used 5 pSR01 colonies and one selection strain colony as a control.
| + | |
| - | *N.B.: this plate we are using is not necessarily meant for OD tests like this and may give some variable readings.
| + | |
| - | '''Lambda red results:'''
| + | |
| - | *Yesterday's lambda red, which used Noah's kan-ZFB-wp-hisura, has yet to produce colonies. We will leave the plates in longer in case any come up later.
| + | |
| - | '''Thio kanZFB:'''
| + | |
| - | *Since yesterday's lambda red may not have worked, we need to get the thio kan up and running. We ran out the touchdown PCR samples (since they have more DNA) and will gel extract.
| + | |
| - | **57.7ng/µL 260/280=1.98
| + | |
| - | **69.3ng/µL 260/280=1.98
| + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | <div id="720" style="display:none">
| + | |
| - | ==July 20th==
| + | |
| - | ===Team ZF===
| + | |
| - | ====Checking on 1/100 ul and 1/10000 ul 78, 79, and 82 plates====
| + | |
| - | We got colonies on all the plates, but the 1/100 ul plates look a bit crowded. On the other hand, the 1/10000 plates only have a few colonies. We decided to save both plates, and decide which to use later.
| + | |
| - | *Based on these results, a 1/1000 ul dilution might be best.
| + | |
| - | | + | |
| - | ====Check on 85 culture====
| + | |
| - | There was no growth in our culture.
| + | |
| - | | + | |
| - | Later: It ends up that our ultramer from 85 (Pos Con 77) had an error in it. Instead of coding for fingers 2 and 3, it codes for fingers 1 and 2. We ordered a new ultramer, and we no longer need to work on 85.
| + | |
| - | | + | |
| - | ====Finalizing the practice plate====
| + | |
| - | We decided on using a practice plate with 24 oligos, consisting of 18 target sequences, and 6 controls. Our 6 controls are Zif268, OZ052, OZ123, and 3 sequences from the CODA table. We will need to construct the expression plasmid for Zif268, OZ052, and OZ123. Their ultramers were ordered today.
| + | |
| - | | + | |
| - | Our 18 targets were GNN/TNN sequences, which are more likely to have a binder, and were distributed among the backbones. The target and control groups each have their own set of primers, to facilitate picking them from the pool.
| + | |
| - | | + | |
| - | ====Expression Plasmid Sequencing Results====
| + | |
| - | We got back our sequencing results:
| + | |
| - | | + | |
| - | {| border="1"
| + | |
| - | | Name||ID||Final?||Notes
| + | |
| - | |-
| + | |
| - | | style="background: green"|'''FH Top (77)'''||1||N||1 snp
| + | |
| - | |-
| + | |
| - | | ||2||N||5 snps
| + | |
| - | |-
| + | |
| - | | || '''3'''||'''Y'''||'''G -> C. Still Ala'''
| + | |
| - | |-
| + | |
| - | | ||4||N||1 snp
| + | |
| - | |-
| + | |
| - | | style="background: red"|'''FH Bottom (78)'''||1||N||1 Deletion at 1746, last nucleotide of F of FQRICMRN
| + | |
| - | |-
| + | |
| - | |style="background: red"| '''CB Top (79)'''||1||N||A lot of deletions/mistakes in f2/f3 region
| + | |
| - | |-
| + | |
| - | |style="background: green"|'''CB Bottom (80)'''||'''1'''||'''Y'''||
| + | |
| - | |-
| + | |
| - | | ||'''2'''||'''Y'''||'''G->C. Stiil Ala'''
| + | |
| - | |-
| + | |
| - | | || 3||N||deletion 1613-1631
| + | |
| - | |-
| + | |
| - | | ||4||N||many mistakes at end, may be ok
| + | |
| - | |-
| + | |
| - | |style="background: green"|'''Myc 198 (81)'''||'''1'''||'''Y'''||
| + | |
| - | |-
| + | |
| - | | ||'''4'''||'''Y'''||'''G->C. Still Ala.'''
| + | |
| - | |-
| + | |
| - | |style="background: green"|'''Myc 981 (82)'''||'''1'''||'''Y'''||
| + | |
| - | |-
| + | |
| - | |style="background: green"|'''Pos Con 16 (83)'''||''' 4'''||'''Y'''||'''G->C. Still Ala.'''
| + | |
| - | |-
| + | |
| - | | ||2||N||A lot of deletions/mistakes in f2/f3 region
| + | |
| - | |-
| + | |
| - | | ||3||N||A lot of deletions/mistakes in f2/f3 region
| + | |
| - | |-
| + | |
| - | |style="background: green"|'''Pos Con 55 (84)'''|| '''1'''||'''Y (?)'''||'''Right sequence, but messy trace'''
| + | |
| - | |-
| + | |
| - | ||| 4||N||at 547: deletion at type II binding site
| + | |
| - | at 665: deletion at beginning of F3
| + | |
| - | |-
| + | |
| - | | style="background: green"|'''Zif 268'''||'''1'''||'''Y'''||
| + | |
| - | |}
| + | |
| - | | + | |
| - | | + | |
| - | {| {{table}}
| + | |
| - | | align="center" style="background:#f0f0f0;"|'''Summary:'''
| + | |
| - | |-
| + | |
| - | | '''Good:''' 77.3, 80.1, 80.2, 81.1, 81.4, 82, 83, 84.1 (?), Zif268
| + | |
| - | |-
| + | |
| - | | '''Bad (Redo):''' 78, 79
| + | |
| - | |}
| + | |
| - | | + | |
| - | We picked 4 new colonies for 78, and 79 since their sequences had many errors. We left these cultures overnight at 37* in the shaker.
| + | |
| - | | + | |
| - | ====Construction of OZ052 and OZ123: Joining the Finger with Homology regions====
| + | |
| - | Previously, we did a PCR to [[Lab_Notebook:_July#Overlap_PCR_of_OZ052_and_OZ123|reassemble the ultramers]] for OZ052 and OZ123, but they only coded strictly for F1/F2/F3 and did not have any overhang homology with any surrounding code. Thus, we performed PCR to add these homology overhangs so that we can integrate them with the omega subunit as well as the spec backbone to create a final expression plasmid.
| + | |
| - | | + | |
| - | The program we used was 55* annealing temperature, and a 15 second extension time. Gel results pictured below.
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.20_OZ_overhang_additions_annotated.png|thumb|Overlap PCR to extend ultramers with homologies]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | We had an expected product size of 400 bp., which we can see in the gel. Bands are a little weird since there are bands heavier than the heaviest theoretical band (which is the strong product). We will do a gel extraction tomorrow to get rid of these bands to obtain pure product for use in the isothermal assembly.
| + | |
| - | | + | |
| - | ===Team TolC===
| + | |
| - | ====rpoZ MAGE knockout====
| + | |
| - | *Ran two rounds of MAGE on 4 ECNR2 cultures with the kan-ZFB-wp insertion using Noah's rpoZ knockout oligo
| + | |
| - | **Grew cultures up to mid-log in kan+amp and LB
| + | |
| - | **Four cultures are ZF091-2d, ZF091-4d, ZF090-1d, and ZF090-5d
| + | |
| - | **Electroporator read 5.4 ms for all four cultures in first round
| + | |
| - | **Could not do a third round of MAGE since the E.Coli do not appear to have recovered, and are nowhere near midlog (MAGE round 2 was done by 5:15pm and the cultures were checked at 8:30pm). Will let the culture go on overnight.
| + | |
| - | **Put 50ul of a 1:100 dilution of each culture on Amp plates. Used a spreader for plates 1&2 and glass beads for plates 3 & 4.
| + | |
| - | | + | |
| - | ====Glycerol Stocks====
| + | |
| - | *Made 4 glycerol stocks of the cultures sent to be sequenced
| + | |
| - | **ZF090-1s, ZF090-2s, ZF090-3s, ZF091-3s
| + | |
| - | ====convinced that the gradient PCR machine is out of order, ought to inform Andrew====
| + | |
| - | | + | |
| - | ===Team Wolfe===
| + | |
| - | '''IPTG Plate and the Plate Reader'''
| + | |
| - | *Technical delays have allowed us only a few hours worth of plate reading for a 46-well plate with selection strain + pSR01 in LB+Amp +/- IPTG, NM+His +/- IPTG, NM +/- IPTG, and NM+3-AT +/- IPTG. Strains used: ∆HisB∆pyrF∆rpoz+pSR01 (5 colonies) and ∆HisB∆pyrF∆rpoz (1 colony).
| + | |
| - | **The plate had a noticeable white residue under the lid around around the edges. We believe this was due to flaking of the plastic cover from vibration in the plate reader.
| + | |
| - | ** Cell growth in the plate suggested that IPTG is killing our cells since few cultures survived with IPTG and those that did (LB+Amp) grew less than the equivalent cultures without IPTG.
| + | |
| - | | + | |
| - | '''New Plate Without IPTG'''
| + | |
| - | * On plate reader at 11:30AM
| + | |
| - | **Strains used: Water as control, Selection (Wolfe) Strain, Selection Strain + pSR01
| + | |
| - | **Media Used: LB+Tet (LB+Amp for SS+pSR01),NM+His, NM, NM+3-AT (1µM), NM+3-AT (2.5µM), NM+3-AT (5µM).
| + | |
| - | | + | |
| - | '''Lambda Red'''
| + | |
| - | Same procedure as usual
| + | |
| - | *Added ~350ng of Thio-Kan-ZFB-wp-His3-Ura3
| + | |
| - | *Used "new cuvettes"
| + | |
| - | *Negative water control
| + | |
| - | | + | |
| - | '''Plate Reader Update'''
| + | |
| - | TEMPORARILY OUT OF SERVICE
| + | |
| - | | + | |
| - | '''Your worst nightmare'''
| + | |
| - | http://www.youtube.com/watch?v=v4o8MDjuu-A
| + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | <div id="721" style="display:none">==July 21st==
| + | |
| - | ===Team ZF===
| + | |
| - | ====Checking colonies 78 and 79====
| + | |
| - | All of the cultures grew overnight, so we performed a miniprep on them to obtain the plasmids.
| + | |
| - | {|
| + | |
| - | |[[File:2011.7.21_Bad_sequence_redo_miniprep.jpg|thumb|Plasmid concentrations after miniprep]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | Our concentrations were better than they have been in the past; notably, after the addition of buffer P2, our solution actually turned a bright blue (indicating lysis of the cells), which we had not seen before.
| + | |
| - | | + | |
| - | We then ran a PCR on our miniprep product, to check for our desired insert. We used our [[#PCR of expression plasmid cross-junction|usual protocol]] for a PCR of the expression plasmid cross-junction.
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.21_New_colonies_junction_pcr_annotated.png|thumb|PCR results, note that some lanes do not show strong bands]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | Some of the products did not show up very strongly, but we sent them out for sequencing in case it was a PCR/gel error.
| + | |
| - | | + | |
| - | ====Sending out our expression colonies for sequencing====
| + | |
| - | We sent out the samples from [[#Expression Plasmid Sequencing Results|yesterday]] that had no more than 1 SNP for reverse sequencing. In addition, we also sent out 78 and 79 for forward and reverse sequencing.
| + | |
| - | *Reverse sequencing: 77.1, 77.3, 77.4, 78.1, 80.1, 80.2, 80.4, 81.1, 81.4, 82.1, 83.4, 84.1, 84.4, Zif268
| + | |
| - | *Forward and reverse sequencing: 781, 78.2, 78.3, 78.4, 79.1, 79.2, 79.3, 79.4
| + | |
| - | | + | |
| - | For sequencing, we used the forward primer pZE23G_3581_F CCAACCTTACCAGAGGGCGC (Junction forward) and the reverse primer pZE23G_2123_R CGAACGACCTACACCGAACT (Junction reverse)
| + | |
| - | | + | |
| - | We did a PCR of the sequences from yesterday that we want sequenced again.
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.21_more_sequencing_pcr_ver_2_annotated.png|thumb|PCR results, looking good]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | The bands were clear, and we will send these products to be sequenced.
| + | |
| - | | + | |
| - | ====Gel Extraction and Isothermal Assembly of OZ fingers+homology====
| + | |
| - | We ran a gel extraction of our PCR product from [[#Construction of OZ052 and OZ123: Joining the Finger with Homology regions|yesterday]], so that we could use it for isothermal assembly later. Strangely, we got bands that were larger than our largest theoretical product.
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.21_OZ_overhang_gel_extraction_annotated.png|thumb|Prepare yourself to be gel-extracted]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | We had the following yields from our gel extraction:
| + | |
| - | ID Conc. (ng/ul) 260/280
| + | |
| - | OZ052 96.3 1.93
| + | |
| - | OZ123 125.5 1.91
| + | |
| - | | + | |
| - | Based on these concentrations, we used the following recipes for our isothermal reaction:
| + | |
| - | *OZ052:
| + | |
| - | **Spec: 0.63 ul
| + | |
| - | **OZ052: 1.56 ul
| + | |
| - | **Omega: 0.13 ul
| + | |
| - | **Water: 1.56 ul
| + | |
| - | **Isothermal assembly solution: 7.5 ul
| + | |
| - | *OZ123:
| + | |
| - | **Spec: 0.63 ul
| + | |
| - | **OZ123: 0.14 ul
| + | |
| - | **Omega: 0.13 ul
| + | |
| - | **Water: 1.60 ul
| + | |
| - | **Isothermal assembly solution: 7.5 ul
| + | |
| - | | + | |
| - | We left the isothermal assembly product in the -20* freezer, and will perform the transformation tomorrow.
| + | |
| - | | + | |
| - | ===Team Tolc===
| + | |
| - | ===Team Wolfe===
| + | |
| - | *Lambda red culture did not grow colonies overnight on a kanamycin plate. This may be due to a low time (4.8ms) during the electroporation.
| + | |
| - | *Repeated lambda red using the same procedure. Got a similar time constant and noticed that the water negative control had more cells than the culture containing the insert, but we went ahead and plated 1mL of each on kan plates.
| + | |
| - | *Also replated the 7/20 lambda red culture to see if we can get any good cells (1.5mL).
| + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | <div id="722" style="display:none">==July 22nd==
| + | |
| - | ===Team ZF===
| + | |
| - | ====Sequencing Results of Expression Plasmids====
| + | |
| - | The good news is that we managed to get a perfect sequence for each of our plasmids. The bad news is that we do not know which sequences are the matches, since the names are all scrambled for some reason (i.e. sample 78.x does not align with FH bottom like it should, but instead aligns perfectly with another plasmid sequence). We will thus need to resend our samples for sequencing to be able to choose the correct plasmids that match. Below is a summary of the scrambled data from the sequencing we sent in yesterday:
| + | |
| - | | + | |
| - | {| {{table}}
| + | |
| - | | align="center" style="background:#f0f0f0;"|'''ID'''
| + | |
| - | | align="center" style="background:#f0f0f0;"|'''Is a perfect match with'''
| + | |
| - | | align="center" style="background:#f0f0f0;"|'''Notes'''
| + | |
| - | |-
| + | |
| - | | Myc 198 (81)||78.2, 78.3||
| + | |
| - | |-
| + | |
| - | | FH Top (77)||81.4, Myc 981||83.4 has 1 deletion
| + | |
| - | |-
| + | |
| - | | CB Bottom (80)||84.4||78.1 has 5 deletions; Zif has 1 deletion
| + | |
| - | |-
| + | |
| - | | FH bottom (78)||n/a||84.1 deletion at beginning of F3
| + | |
| - | |-
| + | |
| - | | CB Top (79)||79.3||
| + | |
| - | |-
| + | |
| - | | Myc 981 (82)||78.4||
| + | |
| - | |-
| + | |
| - | | Zif 268||79.4||
| + | |
| - | |-
| + | |
| - | | Pos Con 55 (83)||79.2||79.3 has deletion at beginning of F3
| + | |
| - | |-
| + | |
| - | | Pos Con 16||79.1||
| + | |
| - | |}
| + | |
| - | | + | |
| - | | + | |
| - | To this end, we redid the PCRs for everything in preparation for the resequencing, using the junction primers and [[#PCR of expression plasmid cross-junction|the previous protocol]]. The resulting gels can be observed below:
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.22_78_and_79_for_sequencing_annotated.png|thumb|E-gel showing bands for samples 78.1-78.4 and 79.1-79.4, expected size of 1.4 kb]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.22_rest_of_pcr_for_sequencing_annotated.png|thumb|Remaining samples, expected size of 1.4 kb]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | From these PCRs, all the reactions look like they produced bands of the right size, a good indicator that the isothermal assemblies worked. Only sequencing will tell whether they are truly the products that we are looking for.
| + | |
| - | | + | |
| - | We sent these samples out for sequencing, making sure that all the samples were in the appropriate tubes.
| + | |
| - | | + | |
| - | ====Transformation for OZ plasmids====
| + | |
| - | Today, we took the results of the isothermal assembly from yesterday and did chemical transformation to introduce the plasmids into the Top 10 ChemComp bacteria, using [[Protocols#Cultures|our protocol for chemical transformations]]. They were left to recover for approximately 1.5 hours. We plated 2 dilutions, 10 ul and 50 ul, and left them overnight at 37* in the incubator.
| + | |
| - | | + | |
| - | ===Team Wolfe===
| + | |
| - | *Yesterday's lambda red plate had several large colonies--but so did the negative control plate. We will go ahead and do a PCR of the 1529620 locus (as previously described) on 12 of the colonies in case one of them happens to have the insert. The colonies were also grown up simultaneously in LB+kan.
| + | |
| - | **Results: all of the colonies were cheaters
| + | |
| - | [[File: 2011.07.22.thiokan insert check1(labeled).ong|thumb|left|1529620 locus PCR 7/22/11]]
| + | |
| - | [[File: 2011.07.22.thiokan insert check2(labeled).png|thumb|none|1529620 locus PCR part 2 7/22/11]]
| + | |
| - | *In case these colonies do not have the insert, as we suspect, we will repeat lambda red with the thio kan-ZFB-wp-his3-ura3 insert
| + | |
| - | **1mL culture for each condition, 210 ng DNA
| + | |
| - | **time constant for thio insert sample was 5.0
| + | |
| - | | + | |
| - | ===Team TolC===
| + | |
| - | ====Gel for Jun's MAGE rpoZ, rowD====
| + | |
| - | *The gel appeared to have bands, so we are running a second PCR to confirm it
| + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | <div id="723" style="display:none">
| + | |
| - | ==July 23rd==
| + | |
| - | ===Team Wolfe===
| + | |
| - | *Lambda red results: no real colonies, only cheaters.
| + | |
| - | </div>
| + | |
| - | | + | |
| - | <div id="724" style="display:none">
| + | |
| - | ==July 24th==
| + | |
| - | ===Team ZF===
| + | |
| - | ====Sequencing Results====
| + | |
| - | We got back our sequencing results from the samples we sent out on Friday, and the results are summarized in the table below. The checkmarks indicate whether the sample is adequate for usage or not.
| + | |
| - | | + | |
| - | {| {{table}}
| + | |
| - | |style="background:#f0f0f0;"|''' '''
| + | |
| - | |style="background:#f0f0f0;"|'''FH top (77)'''
| + | |
| - | |style="background:#f0f0f0;"|'''Notes'''
| + | |
| - | |-
| + | |
| - | | ✓||77.1||Only G -> C in ala
| + | |
| - | |-
| + | |
| - | | ✓||77.3||Only G -> C in ala
| + | |
| - | |-
| + | |
| - | | ||77.4||Single deletion in F3
| + | |
| - | |-
| + | |
| - | | ||||
| + | |
| - | |-
| + | |
| - | |style="background:#f0f0f0;"|''' '''
| + | |
| - | |style="background:#f0f0f0;"|'''FH bottom (78)'''
| + | |
| - | |style="background:#f0f0f0;"|''' '''
| + | |
| - | |-
| + | |
| - | | ||78.1||Many, many SNPs throughout, oddly F and R differ
| + | |
| - | |-
| + | |
| - | | ||78.2||Many, many SNPSs, R starts in a strange place
| + | |
| - | |-
| + | |
| - | | ||78.3||Four deletions throughout on both F and R
| + | |
| - | |-
| + | |
| - | | ||78.4||Many, many SNPs throughout
| + | |
| - | |-
| + | |
| - | | ||Original||Only one SNP in F3, but unacceptable
| + | |
| - | |-
| + | |
| - | | ||||
| + | |
| - | |-
| + | |
| - | |style="background:#f0f0f0;"|''' '''
| + | |
| - | |style="background:#f0f0f0;"|'''CB top (79)'''
| + | |
| - | |style="background:#f0f0f0;"|''' '''
| + | |
| - | |-
| + | |
| - | | ||79.1||Many, many SNPs and deletions throughout, F and R the same
| + | |
| - | |-
| + | |
| - | | ✓||79.2||No SNPs
| + | |
| - | |-
| + | |
| - | | ✓||79.3||No SNPs
| + | |
| - | |-
| + | |
| - | | ||79.4||G -> C in ala, deletion in the Q of the F3 \"FSQ\"
| + | |
| - | |-
| + | |
| - | | ||||
| + | |
| - | |-
| + | |
| - | |style="background:#f0f0f0;"|''' '''
| + | |
| - | |style="background:#f0f0f0;"|'''CB bottom (80)'''
| + | |
| - | |style="background:#f0f0f0;"|''' '''
| + | |
| - | |-
| + | |
| - | | ✓||80.1||No SNPs
| + | |
| - | |-
| + | |
| - | | ||80.2||G -> C in ala, deletion in the homology region shortly after F3 ends
| + | |
| - | |-
| + | |
| - | | ||80.4||G -> C in ala, five deletions in F3/homology region
| + | |
| - | |-
| + | |
| - | | ||||
| + | |
| - | |-
| + | |
| - | |style="background:#f0f0f0;"|''' '''
| + | |
| - | |style="background:#f0f0f0;"|'''Myc 198 (81)'''
| + | |
| - | |style="background:#f0f0f0;"|''' '''
| + | |
| - | |-
| + | |
| - | | ✓||81.1||No SNPs
| + | |
| - | |-
| + | |
| - | | ✓||81.4||Only G -> C in ala
| + | |
| - | |-
| + | |
| - | | ||||
| + | |
| - | |-
| + | |
| - | |style="background:#f0f0f0;"|''' '''
| + | |
| - | |style="background:#f0f0f0;"|'''Myc 981 (82)'''
| + | |
| - | |style="background:#f0f0f0;"|''' '''
| + | |
| - | |-
| + | |
| - | | ✓||82.1||Only G -> C in ala
| + | |
| - | |-
| + | |
| - | | ||||
| + | |
| - | |-
| + | |
| - | |style="background:#f0f0f0;"|''' '''
| + | |
| - | |style="background:#f0f0f0;"|'''Pos con 16 (83)'''
| + | |
| - | |style="background:#f0f0f0;"|''' '''
| + | |
| - | |-
| + | |
| - | | ✓||83.4||Only G -> C in ala
| + | |
| - | |-
| + | |
| - | | ||||
| + | |
| - | |-
| + | |
| - | |style="background:#f0f0f0;"|''' '''
| + | |
| - | |style="background:#f0f0f0;"|'''Pos con 55 (84)'''
| + | |
| - | |style="background:#f0f0f0;"|''' '''
| + | |
| - | |-
| + | |
| - | | ✓||84.1||Only G -> C in ala
| + | |
| - | |-
| + | |
| - | | ||84.4||G -> C in ala, two deletions
| + | |
| - | |-
| + | |
| - | | ||||
| + | |
| - | |-
| + | |
| - | |style="background:#f0f0f0;"|''' '''
| + | |
| - | |style="background:#f0f0f0;"|'''Zif268'''
| + | |
| - | |style="background:#f0f0f0;"|''' '''
| + | |
| - | |-
| + | |
| - | | ✓||Zif268||No SNPs
| + | |
| - | |-
| + | |
| - | |
| + | |
| - | |}
| + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | <div id="725" style="display:none">
| + | |
| - | ==July 25th==
| + | |
| - | ===Team TolC===
| + | |
| - | ====Sequencing of Jun's rpoZ MAGE knockout====
| + | |
| - | *Prepared six products and primers for sequencing
| + | |
| - | **Sending Jun colony 4, PCR's 1-mutant_R, 2-mutant_R, 3-mutant_R, 4-mutant_R, 2-mutant_Int_R, and 4-mutant_Int_R
| + | |
| - | **delayed until tomorrow (July 25, 2011) because pickup for the day already done.
| + | |
| - | | + | |
| - | ====Lambda Red Recombination of zeocine into TolC(assistance from Naomi, Cub team member)====
| + | |
| - | *PCR out zeocin cassette (replacing the rpoZ, for selection) from Wolfe Strain
| + | |
| - | **Used primers rpoZ_F and rpoz_R, expecting a band about 1500bp? Total length about 939bp, including the promoters, according to [[http://rothlab.ucdavis.edu/drugs/zeo.html Roth lab @ UC Davis, Zeocin]]
| + | |
| - | **Ran 8, 25 µL solutions with KAPA
| + | |
| - | **PCR protocol(PCR 7 machine, "ZEOC")
| + | |
| - | ***95˚ C, for 5 min
| + | |
| - | ***98˚ C, for 20 sec
| + | |
| - | ***62˚ C, for 15 sec
| + | |
| - | ***72˚ C, for 45 sec
| + | |
| - | ***Repeat 29 times
| + | |
| - | ***72˚ C, for 5 min
| + | |
| - | ***4˚ C, forever
| + | |
| - | **Gel extracted the PCR product '''NOTE. Since the gel, was so clean, we may do a PCR clean up next time, instead of running the PCR product through a gel for gel extraction. Just make sure to use these exact same parameters.'''
| + | |
| - | ***Nanodrop
| + | |
| - | ****156.2 ng/µL and 107.6 ng/µL, each in 30ul of water (distilled, deionised- molecular grade)
| + | |
| - | *Performed lambda red recombination
| + | |
| - | **Time constant of 5.6 ms
| + | |
| - | | + | |
| - | ====rpoZ MAGE knockout attempt...yet again====
| + | |
| - | *Used new oligo to perform a MAGE knockout of rpoZ
| + | |
| - | **Oligo named rpoZ_MAGE_Subs
| + | |
| - | *Electroporator read 5.2 ms
| + | |
| - | *Allowed for 3 hours of growth and spread on zeocine plate
| + | |
| - | | + | |
| - | ===Team Wolfe Cub===
| + | |
| - | *Plate reader is still being repaired--we will have to wait to check the growth curves
| + | |
| - | *Since plans A and B are not working well at the moment, we will go ahead and start preparing for plan C (using a plasmid). Ordered primers that will clone out the pSB4K5 (a biobrick vector with a kan cassette, http://partsregistry.org/Part:pSB4K5) backbone with homology to the ZFB-wp-his3-ura3 construct. We will then connect the two with Gibson assembly.
| + | |
| - | **To this end, we need to use the biobrick DNA sent to us to transform chemically competent E coli. Unfortunately the 384-well plates that they are stored in could not be found because Andrew is out of town--hopefully they will turn up tomorrow.
| + | |
| - | *For the rest of the day, we joined team TolC for gel extraction, MAGE, and lambda red!
| + | |
| - | | + | |
| - | ===Team ZF===
| + | |
| - | ====Result of OZ052 and OZ123 Transformation====
| + | |
| - | [[#Transformation for OZ plasmids|Last Friday]] we plated out the transformed bacteria for the two OZ controls in 10 ul and 50 ul volumes. They both yielded colonies, but the 10 ul plates only had a handful while the 50 ul plates had around 15-20 colonies each. We picked four colonies from each plate and grew them in LB/spec so that tomorrow we can perform a miniprep and send them off for sequencing. We should know the final verdict by Wednesday regarding which ones are good or bad.
| + | |
| - | | + | |
| - | ====Glycerol stockmaking====
| + | |
| - | We got the sequencing results back from last Friday, and took the following colonies to make into glycerol stock solutions: 77.1, 79.2, 80.1, 81.1 (x2 for both FH bottom and Myc 198), 82.1, 83.4, 84.1, Zif268. These colonies are a subset of the "good" colonies from our [[Lab_Notebook:_July#Sequencing_Results|sequencing results]] from Sunday. We mixed 120 ul of bacteria in mid-log with 300 ul of 80% glycerol to make the final solution, which was stored at -80 degrees C.
| + | |
| - | | + | |
| - | ====Restriction enzyme test====
| + | |
| - | We tested both XbaI and BsaI on the junction PCR product using colony 79.4 as a template (which was used because it was a "bad" sequence overall but had intact BsaI and XbaI cut sites). The junction PCR product is 1326 bp, and we expected to see bands of 593 and 739 bp with XbaI, and 717 and 578 bp with BsaI. Our restriction digest recipe consisted of the following per reaction:
| + | |
| - | *1 ul 10X buffer (NE biolabs buffer 4)
| + | |
| - | *7.9 ul H2O
| + | |
| - | *1 ul DNA (~100 ng)
| + | |
| - | *1 ul enzyme (~2 units)
| + | |
| - | *1 ul BSA (100x)
| + | |
| - | | + | |
| - | The two gels we ran for this digest showed that XbaI did NOT work, which may be due to XbaI being too old. However, we did get double bands both times for BsaI. Our egel was strange because of uneven loading, and so we were unable to determine the true size of both bands, but the agarose gel we ran later showed the two bands in the right positions. The two gels can be seen below:
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.25.enzyme_test_1.png|thumb|E-gel of our first digestion. The gel looks unusual (notice the bubble-like formation), so we performed a second digestion.]]
| + | |
| - | |[[File:2011.7.25_enzyme_test_take2.png|thumb|1% agarose gel of our second digestion. Although the bands are faint, we can see the appropriately sized bands for Bsa1.]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | ====Ultramer Construction of Pos Con 77 and Other F2/F3 positive controls====
| + | |
| - | We redid the ultramer overlap PCR for Pos Con 77, this time with the correct reverse sequence. We also assembled ultramers for Zif268, OZ052 and OZ123 (these are the plasmids that will have the type II cut sites in them along with F2/F3 only, as opposed to having the entire finger already assembled). For the Zif268 reaction, an annealing temperature of 65*C was used, and for the rest, the annealing temperature was 58*C. For all reactions, the extension time was 15 seconds.
| + | |
| - | | + | |
| - | The following primers were used for each reaction (index taken from the Dropbox primer index):
| + | |
| - | *Pos con 77 - 76, 122
| + | |
| - | *Zif268 - 123, 124
| + | |
| - | *OZ052 - 76, 125
| + | |
| - | *OZ123 - 76, 126
| + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | <div id="726" style="display:none">
| + | |
| - | ==July 26==
| + | |
| - | ===Team ZF===
| + | |
| - | ====Isothermal Assembly of Pos Con 77, Zif 268, and OZs====
| + | |
| - | Today we checked the PCR products of the ultramer reaction from yesterday. They did not look good. We repeated the reaction using a touchdown PCR beginning with an annealing temperature of 70 degrees and decreasing 2 degrees every other cycle until hitting a minimum of 58 degrees, and we brought the total number of cycles to 25. Extension time was 30 seconds. This looked great. The pictures for the two reactions can be seen below:
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.26_ultramer_reaction_and_restriction_rerun_annotated.png|thumb|Our first PCR was extremely streaky, so we decided to redo it.]]
| + | |
| - | |[[File:2011.7.26_ultramer_touchdowns_last_four_lanes_annotated.png|thumb|We did a touchdown PCR, and our product looks better, although it's still streaky.]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | Before the isothermal assembly, we did a PCR purification and measured the concentrations of the ultramer reaction products:
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.26_PCR_purification_after_ultramer_assembly.jpg|thumb|Concentrations of our PCR product after PCR purification. This was used in our isothermal assembly.]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | We then performed an isothermal assembly, using the following mix for Pos con 77, OZ052, and OZ123:
| + | |
| - | *0.63 ul of spec backbone at ~157 ng/ul
| + | |
| - | *0.3 ul of ultramer (~50 ng/ul)
| + | |
| - | *0.13 ul of omega subunit/linker
| + | |
| - | *1.44 ul of H2O
| + | |
| - | *7.5 ul of isothermal mix
| + | |
| - | | + | |
| - | For Zif268:
| + | |
| - | *0.63 ul of spec backbone
| + | |
| - | *0.19 ul of Zif (~90 ng/ul)
| + | |
| - | *0.13 ul of omega/linker
| + | |
| - | *1.55 of H2O
| + | |
| - | *7.5 ul of isothermal mix
| + | |
| - | | + | |
| - | This was left at 50 degrees for an hour, then stored at 4 degrees.
| + | |
| - | | + | |
| - | ====Prepare control OZs for sequencing====
| + | |
| - | We performed a miniprep on the eight samples for sequencing, obtaining the following concentrations:
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.26_Miniprep.jpg|thumb|Our OZ concentrations after miniprep.]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | The miniprep lysis step looked good, and the samples turned blue after lysis.
| + | |
| - | | + | |
| - | We then performed a junction PCR using the [[Lab_Notebook:_July#PCR_of_expression_plasmid_cross-junction|previous protocol]], and checked the products on a gel:
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.26_oz052_and_oz123_for_seq_annotated.png|thumb|Gel of our PCR product that we sent out for sequencing.]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | They looked good, so we sent them out for sequencing. Hopefully we will have our final OZ control plasmids for tomorrow.
| + | |
| - | | + | |
| - | ====Practice Plate resuspension====
| + | |
| - | We resuspended the practice plate oligos in 40 ul of water to produce a 100 uM solution, taking 10 ul of the final solutions into 90 ul of water in separate tubes to create 10 uM working concentrations. When we receive our tag primers, we should be able to test the presence of the oligos in each tube as well as amplify them in preparation for integration into the expression plasmids.
| + | |
| - | | + | |
| - | ====Colony finalization====
| + | |
| - | For the colonies that we plated yesterday, we saw that the plates all had good growth on them and so we wrapped the plates in parafilm and put them at 4 degrees for storage. 1/1000 ul from saturated culture seems to be a good amount to plate.
| + | |
| - | | + | |
| - | ===Team TolC===
| + | |
| - | ====Zeocin insertion into ECNR2====
| + | |
| - | *Plated 3 mL of our recombination culture on zeocin plate, and also plated 2 mL of MAGE results on zeocin plate as a control
| + | |
| - | **The recombination plate had about 15 colonies around 9 30am, and the MAGE plate had none
| + | |
| - | *Continued by PCR of 4 colonies on recombination plate
| + | |
| - | *Each template was analysed with presence of different sized bands for rpoZ (using rpoZ_F and rpoZ_R) and presence of Zeocin cassette (using rpoZ_R and zeocin_R)
| + | |
| - | *Gel resulted with the following:
| + | |
| - | [[File: 2011.07.26.Zeocininsertion(labeled).png|thumb|none|Zeocin insert check 7/26/11]]
| + | |
| - | | + | |
| - | **'''SUCCESS!!! Yippee!!!'''
| + | |
| - | **NOTE. Since there was a bit of non specific annealing, we might want to use a higher annealing temperature of 64C instead of 62C
| + | |
| - | *Decided to run PCR for the Kan insertion just to make sure it was still present in the 4 colonies we chose, and it is!!!
| + | |
| - | [[File: 2011.07.26.kaninsertioncheck(labeled).png|thumb|none|Kan-ZFB-wp insert check 7/26/11]]
| + | |
| - | | + | |
| - | WERE READY TO ROOOCCKKK!!!!!
| + | |
| - | | + | |
| - | ====MAGE rpoZ knockout====
| + | |
| - | *Plated 1:10 and 1:100 dilution of MAGE results, and this morning at 10 am, they both had cells and in appropriate proportions
| + | |
| - | **If above PCR and gel of zeocin insertion does not appear to be successful, then we will run 50 PCR on 50 different colonies on the 1:100 dilution plate of MAGE rpoZ knockout
| + | |
| - | | + | |
| - | ===Team Wolfe===
| + | |
| - | *Plate reader: trying to get in contact with Christian Daly who runs the Spectramax plate reader in the Bauer facility (Northwest basement). Hopefully we will get access soon.
| + | |
| - | *Selection system plasmid: still trying to get hold of the biobricks. Designed Quikchange primers to change the zinc finger binding sites.
| + | |
| - | *Lambda red recombination: gave lambda red one last chance, this time with a positive control
| + | |
| - | **negative control (water): plated 1mL
| + | |
| - | **positive control (kan cassette from Wyss): 50ng DNA, plated 1mL and 100uL
| + | |
| - | **thio kan-ZFB-wp-his3-ura3: 180ng DNA, plated 1mL
| + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | <div id="727" style="display:none">
| + | |
| - | ==July 27th==
| + | |
| - | *Impromptu meeting for all teams, TFs, and advisors in NW labs basement, to discuss "how we will move forward to test the selection, now that we have a TolC strain that should work for selection and a His3Ura3 plasmid-based selection that is almost complete," and to lay out and assign a lab to-do for iGEM parts, selection systems, and what to do in preparation for The Chip to arrive/backup plan in case it arrives too late (see notes in the dropbox under "Selection Meeting 7-27-11.docx")
| + | |
| - | ===Team TolC===
| + | |
| - | ====Culture of TolC+Kan-ZFB-wp+Zeocin====
| + | |
| - | *Did not realize that colonies needed to be grown up without antibiotic before being grown in Kanamycin, so all the three colonies from yesterday passed away and didn't make it through the night :(, so we grew up 4 more colonies and ran PCR and gel to solidify the presence of the necessary insertions
| + | |
| - | | + | |
| - | ===Team ZF===
| + | |
| - | ====Checking the OZ sequencing results====
| + | |
| - | {| {{table}}
| + | |
| - | | align="center" style="background:#f0f0f0;"|''' '''
| + | |
| - | | align="center" style="background:#f0f0f0;"|'''OZ052'''
| + | |
| - | | align="center" style="background:#f0f0f0;"|'''Notes'''
| + | |
| - | |-
| + | |
| - | | ✓||1||No snps
| + | |
| - | |-
| + | |
| - | | ✓||2||No snps
| + | |
| - | |-
| + | |
| - | | ✓||3||No snps
| + | |
| - | |-
| + | |
| - | | ||4||Deletion at F1
| + | |
| - | |-
| + | |
| - | | align="center" style="background:#f0f0f0;"|''' '''
| + | |
| - | | align="center" style="background:#f0f0f0;"|'''OZ123'''
| + | |
| - | | align="center" style="background:#f0f0f0;"|'''Notes'''
| + | |
| - | |-
| + | |
| - | | ✓||1||No snps
| + | |
| - | |-
| + | |
| - | | ||2||deletion in omega linker
| + | |
| - | |-
| + | |
| - | | ✓||3||No snps
| + | |
| - | |-
| + | |
| - | | ✓||4||No snps
| + | |
| - | |}
| + | |
| - | | + | |
| - | We made glycerol stocks and final plates from 0Z052.1 and OZ123.1.
| + | |
| - | | + | |
| - | ====Transformation of Pos Con 77, OZs, Zif 268====
| + | |
| - | We transformed Top 10 One Shot Electrocomp bacteria with our isothermal assembly product from yesterday. We let the cells recover for an hour, and plated 50 ul, leaving them at 37* overnight.
| + | |
| - | | + | |
| - | ===Team Wolfe===
| + | |
| - | | + | |
| - | *Morning meeting: Since TolC has gotten their selection system onto the genome, our priorities are now going to be making the plasmid-based Wolfe selection system and getting data on the plate reader. We also will eventually try to use a genome-based selection system but using the EcNR2 strain (oligos have been ordered)
| + | |
| - | '''Plate Reader:'''
| + | |
| - | *We took a trip downstairs to B2 and were trained to use the Bioscreen plate reader in the Bauer Core facility. We now have access to a working plate reader!
| + | |
| - | *Bioscreen uses a 100-well honeycomb plate, so the organization was as follows:
| + | |
| - | **Well 1, 11, 21, etc: LB+tet
| + | |
| - | **Well 2, 12, 22, etc: NM+his
| + | |
| - | **Well 3, 13, 23, etc: NM
| + | |
| - | **Well 4, 14, 24, etc: NM+1mM 3-AT
| + | |
| - | **Well 5, 15, 25, etc: NM+2.5mM 3-AT
| + | |
| - | **Well 6, 16, 26, etc: NM+5mM 3-AT
| + | |
| - | **Well 7, 17, 27, etc: LB+tet+100µM IPTG
| + | |
| - | **Well 8, 18, 28, etc: NM+IPTG
| + | |
| - | **Well 9, 19, 29, etc: NM+1mM 3-AT+IPTG
| + | |
| - | **Well 10, 20, 30, etc: NM+5mM 3-AT+IPTG
| + | |
| - | **Column 1: no cells
| + | |
| - | **Column 2-3: ∆HisB∆pyrF∆rpoZ
| + | |
| - | **Columns 4-10: ∆HisB∆pyrF∆rpoZ+pSR01
| + | |
| - | '''Electrocomp Transformation'''
| + | |
| - | *Used Team ZF's protocol to transform electrocompetent cells with two of the pSB4K5 plasmids found in our biobricks plates (2:17H and 3:11P)
| + | |
| - | *Used 10µL ddH2O to resuspend DNA in biobrick plate wells and transferred to eppendorf.
| + | |
| - | *2µL DNA (which, interestingly, was red...)
| + | |
| - | *plated 10µL and 100µL of each on kan plates
| + | |
| - | '''Quikchange primers:'''
| + | |
| - | *Realized that we may need to consider the bases on either side of the zinc finger binding site, so we will check this and put off ordering another day.
| + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | <div id="728" style="display:none">==July 28th==
| + | |
| - | ===Team Wolfe===
| + | |
| - | '''Plate reader and pSR01:'''
| + | |
| - | *the Bauer plate reader seems to have worked very well, bu† unfortunately none of our pSR01-transformed colonies grew. This is probably because the plates were a couple weeks old. Unfortunately our glycerol stock of the strain also is missing, so we will try growing up some of the colonies in case on of them works. Otherwise, we will grow up ∆HisB∆pyrF∆rpoZ and transform again.
| + | |
| - | *Transformation: usual protocol, about 100µL DNA, recovery 2hrs, plate 1µL, 10µL, 100µL.
| + | |
| - | '''pSB4K5:'''
| + | |
| - | *The kan plates from the transformation all had colonies! We chose one from 2:17H and one from 3:11P to grow up in LB+kan for miniprep and glycerol stocks.
| + | |
| - | *Ordered quikchange primers: for OZ052 and OZ123 we added the context bases 5' and 3' of the binding sequence (found in OPEN supplementary materials). Since CODA did not include the extra bases, we left them the same as the original plasmid (i.e. Zif268's).
| + | |
| - | | + | |
| - | ===Team TolC===
| + | |
| - | ====cultures of ECNR2+Kan-ZFB-wp+zeocin from F9,F11, for glycerol stocks and transformation====
| + | |
| - | ====Sequencing====
| + | |
| - | *Ran PCR and sent out sequencing to solidify the presence of the Kan+ZFB+wp and zeocin within the ECNR2 genome
| + | |
| - | **Used rpoZ_R, rpoZ_F for PCR of the zeocin cassette (and rpoZ_F for sequencing) and TolC_Seq_R, TolC_Seq_F for PCR of the Kan-ZFB-wp insert (and tolC_seq_F for sequencing)
| + | |
| - | | + | |
| - | ====Plasmid transformation====
| + | |
| - | *Performed transformation on the Zif268 plasmid into the ECNR2 culture
| + | |
| - | **Used 3 µL in a 50 µL dilution (49+1) of 1 ul of 55 ng/µL concentration of plasmids
| + | |
| - | **1.82 V for 5.00 ms according to the electroporator
| + | |
| - | | + | |
| - | ===Team ZF===
| + | |
| - | ====Checking on Pos Con 77, OZs, and Zif 268 plates====
| + | |
| - | Unfortunately, nothing grew on our plates from yesterday. Our time constants from the electroporation were rather low (~3.8), so 50 ul may not be enough to see growth. We decided to spin down the cells, resuspend them in 50 ul of LB, and re-plated this solution. We also re-did the transformation using ChemComp cells, and made 100 ul plates. Both sets of plates were left overnight at 37*.
| + | |
| - | | + | |
| - | ====Designing the reporter strain====
| + | |
| - | Instead of placing the GFP reporter on a separate plasmid, we decided to insert it on our expression plasmid, immediately after our zinc fingers, and before the transcription terminator. We placed a ribosome binding site (Shine Dalgarno sequence: AGGAGGTT before the GFP. We will only add the GFP to Zif 268 (whole) because if we can show through GFP that the selection system works for Zif 268, we can assume it also works for the other zinc fingers. This is easier and more cost effective than ordering 12 different primers for each of our plasmids.
| + | |
| - | | + | |
| - | We will construct the reporter strain by performing a PCR to get the plasmid in linear form. We will also perform a second PCR to get out GFP and add homology regions to our Zif 268 plasmid, as well as adding the Shine-Dalgarno sequence. The primers for these PCRs were ordered today. We'll then assemble these pieces to construct a whole plasmid, and transform our bacteria.
| + | |
| - | | + | |
| - | ====PCR with Practice Plate====
| + | |
| - | We performed a PCR to amplify our F1s from the practice plate. We used our primers for CB bottom (primer index: 66 and 67) for the 18 targets and our primers for the controls (primer index: 72 and 73) for the 6 controls. We had an annealing temperature of 52 degrees and an extension time of 15 seconds.
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.28_practice_plate_primer_test_annotated.png|thumb|Practice plate primer test]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | We got products for all of the oligos at our expected product size of 131 bp.
| + | |
| - | | + | |
| - | ====Glycerol stocks of successful plasmids====
| + | |
| - | We happened to check our glycerol stocks, and noticed that the glycerol and bacteria were not mixed thoroughly. We remade all of our glycerol stocks and put them into the -80*.
| + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | <div id="729" style="display:none">
| + | |
| - | ==July 29th==
| + | |
| - | ===Team Wolfe===
| + | |
| - | '''pSR01:'''
| + | |
| - | *The colonies in the 96 well plate did grow overnight, but they were not saturated, implying that they were very unhealthy. We will PCR a few of them (1,2,3,6) just to see if they actually have the plasmid still
| + | |
| - | *Transformation plates look great--we will PCR 12 colonies to verify the plasmid is present
| + | |
| - | *PCR: colonies/culture diluted 1:20 as template (and colonies grown up simultaneously); M13_F and M13_R primers; 50˚C annealing and 3min elongation; 25 cycles
| + | |
| - | *Results: bands are similar to the ones seen the last time we did this PCR (and those samples had also been sequence-verified). These colonies should be good to go!
| + | |
| - | [[File: 2011.07.29.pSR01check(labeled).png|thumb|none|pSR01 check 7/29/11]]
| + | |
| - | '''pSB4K5:'''
| + | |
| - | *Miniprepped both cultures. Yields weren't great (in the case of 2:17H, there actually was a spill) but they are still usable.
| + | |
| - | **2:17H: 6.4 ng/µL 260/280=2.2
| + | |
| - | **3:11P: 9.6ng/µL 260/280=2.4
| + | |
| - | *PCR to add ZFB-wp-his3-ura3 homology:
| + | |
| - | **1µL of plasmid as template
| + | |
| - | **pSB4K5_F+ZFBhl and pSB4K5_R+URA3hl primers
| + | |
| - | **58˚C anneal, 3 min elongation, 25 cycles
| + | |
| - | *Results: the product was really really really faint--primer dimers were by far the bulk of the product. We realized this is probably because one of the primers includes a Not1 restriction site, which is palindromic, 8 bases long, and all Cs and Gs.
| + | |
| - | [[File: 2011.07.29.pSB4K5(labeled).png|thumb|none|pSB4K5 PCR to add homology 7/29/11]]
| + | |
| - | *In order to counteract the primer dimers, we will try a gradient PCR with a range of higher annealing temperatures:
| + | |
| - | **same protocol as before except the annealing temp went from 58-65 with 8 samples (1=58, 8=65)
| + | |
| - | **Results: 3:11P did have visible product bands, but the primer dimers are still stronger
| + | |
| - | [[File: 2011.07.29 pSB4K5 grad(labeled).png|thumb|none|pSB4K5 gradient PCR 7/29/11]]
| + | |
| - | | + | |
| - | ===Team TolC===
| + | |
| - | ====Sequencing of Kan+ZFB+wp and Zeocin insertions====
| + | |
| - | *Sequencing results were successful and solidified presence of both insertions
| + | |
| - | ====Zif268 Plasmid Insertion into ECNR2+Kan-ZFB-wp+zeocin====
| + | |
| - | *The plates had growth on them!!!
| + | |
| - | *So we chose 5 colonies and ran PCR on them to gel them out
| + | |
| - | **The gel shows success for colonies 2,3,4, and 5 that we chose!!!
| + | |
| - | [[File: 2011.07.29.zif268plasmidcheck1.png|thumb|none|Zif268 plasmid insertion check 7/29/11]]
| + | |
| - | | + | |
| - | '''READY FOR SELECTION!!!'''
| + | |
| - | | + | |
| - | ===Team ZF===
| + | |
| - | ====Transformation plating results====
| + | |
| - | We got no growth on any of the plated transformed bacteria for either the ElectroComp or ChemComp bacteria. To check whether the isothermal assembly itself was the problem, we ran a junction PCR using our [[Lab_Notebook:_July#PCR_of_expression_plasmid_cross-junction|previous protocol]] on the isothermal assembly mix, along with a positive control (Myc981):
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.29_isothermal_assembly_junction_test_annotated.png|thumb|No bands visible for the isothermal assembly]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | As observed, there were no bands in the isothermal mix lanes while the positive control worked. Thus, we can conclude that our isothermal assembly did not work, and we will need to repeat it again (currently waiting for more isothermal mix to arrive from Wyss).
| + | |
| - | | + | |
| - | ====Amplification and purification of practice plate oligos====
| + | |
| - | In preparation for our swap reactions (switching the oligo F1s into our expression plasmid), we mapped out the following two experiments that we will need to perform:
| + | |
| - | *Testing the selection system by preparing our expression plasmids in parallel (i.e. creating 18 plasmids in separate reactions), and then combining them all at the end and transforming them into the selection strains. This will allow us to cut and ligate the plasmids without much concern, but test whether we will be able to pick out the binder amongst the rest of the non-binders.
| + | |
| - | *Testing the cutting/ligation by combining all of the oligos first, and then carrying out the construction of the different expression plasmids all in the same tube.
| + | |
| - | | + | |
| - | Note that the results for both will ultimately yield DNA libraries if they work (except that the first experiment is much more foolproof in terms of actually yielding a library than the second experiment).
| + | |
| - | | + | |
| - | To carry out the first experiment, the following steps need to be undertaken:
| + | |
| - | #Perform PCRs on each of the practice plate oligos such that we end up with a relatively high concentration of DNA after PCR and PCR purification (over 40 ng/ul).
| + | |
| - | #Cut both plasmid backbone and oligos with BsaI type II restriction enzyme.
| + | |
| - | #PCR purify restriction products to get rid of enzyme and junk DNA.
| + | |
| - | #Run phosphatase reaction to prevent autoligation.
| + | |
| - | #Mix and ligate to get full plasmid.
| + | |
| - | #*Some tips:
| + | |
| - | #*Each ligation reaction should only have 10-15 ul volume.
| + | |
| - | #*You should have 100 ng of backbone per reaction
| + | |
| - | #**Since we are planning 18 reactions, we will need 1800 ng of backbone, which is several minipreps worth
| + | |
| - | #*The backbone:oligo ratio should be appoximately 1:4
| + | |
| - | #**In our case, the backbone is 2.2 kb and the insert is 131 bp, so for 100 ng of backbone there should be ~20 ng of insert
| + | |
| - | #Gel extract plasmid to get final library.
| + | |
| - | | + | |
| - | Today, we did PCRs and PCR purifications for step one as outlined above. We tested the concentrations of the results, displayed below:
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.29_Practice_Plate_oligo_pcr_pure.jpg|thumb|DNA concentrations]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | In order to have enough backbone to work with, we seeded some LB/spec with CB bottom bacteria in preparation for 4 minipreps of 3 mL each tomorrow. This should definitely yield the 1800 ng of backbone we will need for the ligation.
| + | |
| - | | + | |
| - | ====Practice plate primer tag test====
| + | |
| - | We tested the primers to see if their selection of subpools was exclusive enough or not (i.e. to make sure that our control primers did not amplify the CB bottom sequences, etc.). We ran a PCR crossing primers with DNA with the following results:
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.29_practice_plate_neg_controls_annotated.png|thumb|Strange results]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | The first and third lanes are CB primer/CB DNA and control primer/control DNA respectively, which should yield bands. Lanes two and four were control primer/CB DNA and CB primer/control DNA respectively, which should yield no bands. The fact that we got bands for these lanes is a little bit worrying, and so we are repeating this PCR again to see if we get the same results when we run the gel tomorrow.
| + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | <div id="730" style="display:none">
| + | |
| - | ==July 30th==
| + | |
| - | ===Team TolC===
| + | |
| - | *Checked the plate with ECNR2 with Zeocin knocked into the place of rpoZ. There are appear to be some 15-20 colonies on the plate. This strain is ready for transformation with the zif268 plasmid
| + | |
| - | ===Team ZF===
| + | |
| - | ====CB bottom miniprep====
| + | |
| - | Today we performed a miniprep on the 4 tubes of CB bottom bacteria with 4 ml of culture in each tube. 6 ml of culture were eventually consolidated into a single spin tube during the miniprep, so this would be the equivalent of 4 minipreps in one tube. Of note, after lysis (during addition of buffer P2), the tubes turned extremely blue, darker blue than we had ever seen before. This would seem to indicate that the lysis step occurred very successfully. However, even after consolidating four minipreps into one tube, we still got terrible concentration results for our samples, displayed below:
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.30_CB_bottom_miniprep.jpg|thumb|DNA concentrations after miniprep]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | We will thus seed more LB/spec culture with CB bottom bacteria tomorrow, and perform the miniprep on Monday with TF supervision to hopefully troubleshoot some of the problems with low yield.
| + | |
| - | | + | |
| - | Also of note is that the bacteria were left on the shaker for approximately 23 hours at the time of miniprep, which is longer than the ideal 12-14 hours.
| + | |
| - | </div>
| + | |
| - | | + | |
| - | | + | |
| - | <div id="731" style="display:none">==July 31st==
| + | |
| - | ===Team ZF===
| + | |
| - | ====Primer tag test two====
| + | |
| - | On Friday, we repeated the PCR to test for sub-pool specificity of our primer oligo tags for the chip, using the same PCR protocol. Today, we ran a gel to see if the results were any different, but alas, they were the same. It seems that the primers for CB bottom also amplify out the control sequences, which means that our primer tags are not as specific as we thought they were. The gel can be seen below, with the lanes arranged as follows: CB primer + CB DNA, control primer + CB DNA, control primer + control DNA, and control primer + CB DNA. We would ideally expect bands in lanes 1 and 3 and no bands in lanes 2 and 4. However, this was not the case:
| + | |
| - | | + | |
| - | {|
| + | |
| - | |[[File:2011.7.31_primer_tag_test_two_annotated.png|thumb|PCR repeat results]]
| + | |
| - | |}
| + | |
| - | | + | |
| - | We might try doing touchdown PCR or raising the annealing temperature to get more specificity in the future. It may also be worth ordering oligos to represent the other 5 subpools of targets and cross test the primers with each DNA subpool to make sure that the primers are specific enough to pull out only one subpool. In the end though, this specificity is not critical to our success, and so it is ok if it doesn't eventually work.
| + | |
| - | | + | |
| - | ====Preparing for another CB bottom miniprep====
| + | |
| - | Today we seeded 4 tubes of culture with 100 ul of CB bottom bacteria at saturation. Each tube carried 4 ml of culture. The tubes were put on the shaker at approximately 6:30 pm, so they will be ready for miniprep tomorrow morning after the lab meeting.
| + | |
| - | </div> | + | |
 "
"