Team:Penn State/Project
From 2011.igem.org
(→Overall project) |
(→RecA Co-Protease) |
||
| (58 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| - | |||
| - | |||
<html> | <html> | ||
| - | < | + | <head> |
| - | < | + | <style type="text/css"> |
| - | + | body {background-color:MidnightBlue;} | |
| - | + | </style> | |
| - | + | </head> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | </ | + | |
</html> | </html> | ||
| - | < | + | ==Abstract== |
| + | <center> [[Image:RadioColiTitle.png|600px]]</center> | ||
| - | + | Ionizing radiation and radiation pollution is an important environmental problem that not only affects those working around radiation facilities, but those dealing with the aftermath of widespread nuclear disasters such as those at the Fukushima Daiichi nuclear reactor or the Chernobyl reactor. Penn State’s team project will focus on using a genetic circuit introduced into E. coli bacterial cells, in order to rapidly detect and report the presence of harmful ionizing radiation. We are working to develop a robust and reliable biosensor which utilizes the lambda phage lytic-lysogenic switch coupled with a fast-acting reporter capable of producing an easily visible effect. We believe that the final construct may have the potential to rival current radiation detection methods, such as digital dosimeters. | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | Our hope is that the basis of our biological dosimeter system will prove to be an effective genetic system capable of detecting harmful levels of radiation and relaying it to those working in the field or affected area. We envision our system not only being useful in such applications, but also being capable of further expansion and evolution through the expanding field of synthetic biology. | |
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | == '''Overall project: Bacterial Dosimeter''' == | ||
| + | Radiation of one form or another is a constant presence throughout our every day lives. Radiation, or the transmission and absorption of energy over a given distance, has proven an invaluable technology useful in applications for everything from heating our food to diagnosing and treating diseases. Some forms of radiation, however, can be detrimental to the human body, as they are a common cause of such diseases as radiation poisoning and cancer. | ||
| - | + | All types of radiation can be divided into two categories: non-ionizing and ionizing. Non-ionizing radiation (consisting of radio, micro, infrared, and visible electromagnetic waves) contains less energy and has a relatively small effect on living organisms that has only recently been studied. Ionizing radiation, however, contains a much greater amount of energy capable of ionizing atoms which can lead to harmful effects on living tissue. This category of radiation encompasses alpha and beta decay as well as neutron, X-ray, and gamma radiation. | |
| - | + | ||
| - | + | ||
| - | |||
| - | < | + | <center>[[Image:Electromagnetic-spectrum.jpg]]</center> |
| - | |||
| - | < | + | Our project focuses on detecting the degradation and damage of DNA associated with ionizing radiation. The initial proposed design is shown below. It consists of two parts: a sensor based on a lambda phage bistable switch, and a fast-acting reporter similar to the reporter designed by the Imperial College of London 2010 iGEM team. |
| - | </ | + | |
| + | <center>[[File:PSURad.png|850px|]]</center> | ||
== Project Details== | == Project Details== | ||
| + | ===The Sensor=== | ||
| + | The design of our sensor was based on the lambda phage lysogenic vs lytic switch. Optimally, the system would be activated in the presence of DNA damage due to radiation. For that reason, we utilized the lambda phage switch, which transitions from the lysogenic cycle to the lytic cycle when DNA damage is detected. | ||
| + | The genetic circuit we designed focuses on the DNA damage-sensitive lambda phage lytic-lysogenic cycle switch followed by a rapid response reporter similar to the immobilized fusion enzyme system pioneered by the Imperial College of London 2010 iGEM team. An initial design of our sensor circuit is illustrated below. | ||
| + | <center>[[Image:Sensor1.jpg]]</center> | ||
| - | + | Under normal conditions, PRM is active, which transcribes the lambda repressor, cI + RBS. The lambda repressor binds to OR1 and OR2, repressing PR, which in turn inhibits the transcribtion of Cro and TEV. | |
| + | <center>[[Image:Sensor2.jpg|700px|]]</center> | ||
| + | Once DNA damaged occurs, the ssDNA binds to RecA. RecA becomes activated and cleaves the cI repressor, allowing for PR to turn on. Cro then binds to OR3, which represses PRM and inhibits the transcription of the cI repressor. Now that PR is active, the transcription of the TEV protease occurs, which is used in the reporter structure of the circuit. | ||
| + | === The Reporter === | ||
| + | |||
| + | |||
| + | {| width="800px" | ||
| + | |- | ||
| + | |Our reporter system will use a modified version of Imperial College London iGEM 2010 fast reporter system. Imperial’s quick acting response module used the transcription of TEV protease to cleave the bond between the GFP, C23O fusion protein. This unbound form of C230 could then form a tetramer and convert the colorless substrate catechol into a yellow product. | ||
| + | |[[Image:oldreporter.png|align="center"]] | ||
| + | |} | ||
| + | |||
| + | |||
| + | {| width="800px" vertical-align="top" | ||
| + | |- | ||
| + | |Using the Freiburg BioBricking Assembly technique, we plan to produce a miniature catalog of fast acting reporters while reducing the leakiness of Imperial’s construct. To achieve the latter, we plan to link a second GFP to the C-terminus of C230; this new linkage would be cleavable by RecA. | ||
| + | |[[Image:PSUreporter.png|align="center"]] | ||
| + | |} | ||
| + | |||
| + | <center>[[Image:ReporterTable.png|620px|]]</center> | ||
| + | |||
| + | === RecA Co-Protease === | ||
| + | |||
| + | The normal laboratory strain of E. coli, DH10B, contains an inactivated form of RecA known as RecA1. Therefore, a series of mutations must occur in order to restore its proteatic function and reduce recombination. | ||
| + | |||
| + | '''Activation of RecA''' | ||
| + | ::*Point Mutation at b.p 720: A ⇒ G | ||
| + | '''Removal of Recombinase Activity''' | ||
| + | ::*Arginine 243 ⇒ Glutamine | ||
| + | ::*Lysine 286 ⇒ Asparagine | ||
| + | |||
| + | In order to use the standard bio-bricking techniques, we also had to remove these naturally occurring enzymatic restriction sites | ||
| + | *'''PST1 Site''' [CTGCAG] | ||
| + | **CAG ⇒ CTG (Codon usage bias decreases: .69 to .31) | ||
| + | *'''EcoR1 Site''' [GAATTC] | ||
| + | **TTC ⇒ TTT (Codon usage bias increases: .49 to .51) | ||
| - | |||
| + | '''RecA Test Circuit:''' The cI repressor is constitutively produced and binds to the repressible promoter under normal settings.[[File:RecA Test Circuit1.png|center|]] | ||
| + | Activated RecA then cleaves the cI repressor, causing transcription of RFP. However, if RecA exhibits recombinant activity, then the homologous double terminator regions will recombine. This causes the deletion of the repressible promoter and RFP. | ||
| + | [[File:RecATest2.png|center|]] | ||
=== Part 3 === | === Part 3 === | ||
| Line 82: | Line 94: | ||
== Results == | == Results == | ||
| + | |||
| + | {| style="border-collapse:collapse; color:#FFFFFF;background-color:#000066;" cellpadding="3" cellspacing="2" border="1" bordercolor="#ffffff" width="65%" align="center" | ||
| + | !align="center"|[[Team:Penn_State|Home]] | ||
| + | !align="center"|[[Team:Penn_State/Team|<span style="color:#FFFFFF;">Team</span>]] | ||
| + | !align="center"|[https://igem.org/Team.cgi?year=2011&team_<span style="color:#FFFFFF;">Penn State Official Team Profile</span>] | ||
| + | !align="center"|[[Team:Penn_State/Project|<span style="color:#FFFFFF;">Project</span>]] | ||
| + | !align="center"|[[Team:Penn_State/Parts|<span style="color:#FFFFFF;">Parts Submitted to the Registry</span>]] | ||
| + | !align="center"|[[Team:Penn_State/Modeling|<span style="color:#FFFFFF;">Modeling</span>]] | ||
| + | !align="center"|[[Team:Penn_State/Notebook|<span style="color:#FFFFFF;">Notebook</span>]] | ||
| + | !align="center"|[[Team:Penn_State/Safety|<span style="color:#FFFFFF;">Safety</span>]] | ||
| + | !align="center"|[[Team:Penn_State/Attributions|<span style="color:#FFFFFF;">Attributions</span>]] | ||
| + | |} | ||
Latest revision as of 14:10, 15 July 2011
Contents |
Abstract

Ionizing radiation and radiation pollution is an important environmental problem that not only affects those working around radiation facilities, but those dealing with the aftermath of widespread nuclear disasters such as those at the Fukushima Daiichi nuclear reactor or the Chernobyl reactor. Penn State’s team project will focus on using a genetic circuit introduced into E. coli bacterial cells, in order to rapidly detect and report the presence of harmful ionizing radiation. We are working to develop a robust and reliable biosensor which utilizes the lambda phage lytic-lysogenic switch coupled with a fast-acting reporter capable of producing an easily visible effect. We believe that the final construct may have the potential to rival current radiation detection methods, such as digital dosimeters.
Our hope is that the basis of our biological dosimeter system will prove to be an effective genetic system capable of detecting harmful levels of radiation and relaying it to those working in the field or affected area. We envision our system not only being useful in such applications, but also being capable of further expansion and evolution through the expanding field of synthetic biology.
Overall project: Bacterial Dosimeter
Radiation of one form or another is a constant presence throughout our every day lives. Radiation, or the transmission and absorption of energy over a given distance, has proven an invaluable technology useful in applications for everything from heating our food to diagnosing and treating diseases. Some forms of radiation, however, can be detrimental to the human body, as they are a common cause of such diseases as radiation poisoning and cancer.
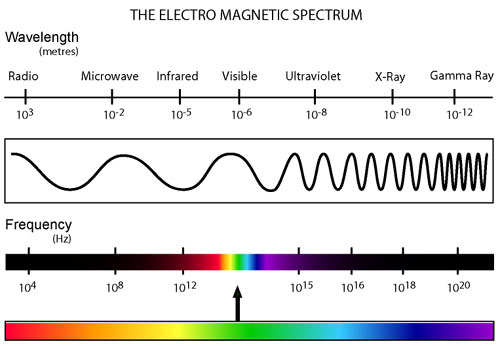
All types of radiation can be divided into two categories: non-ionizing and ionizing. Non-ionizing radiation (consisting of radio, micro, infrared, and visible electromagnetic waves) contains less energy and has a relatively small effect on living organisms that has only recently been studied. Ionizing radiation, however, contains a much greater amount of energy capable of ionizing atoms which can lead to harmful effects on living tissue. This category of radiation encompasses alpha and beta decay as well as neutron, X-ray, and gamma radiation.
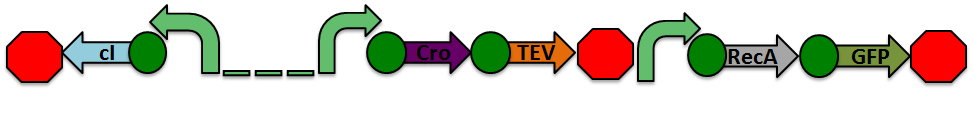
Our project focuses on detecting the degradation and damage of DNA associated with ionizing radiation. The initial proposed design is shown below. It consists of two parts: a sensor based on a lambda phage bistable switch, and a fast-acting reporter similar to the reporter designed by the Imperial College of London 2010 iGEM team.
Project Details
The Sensor
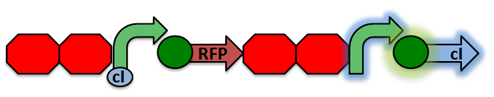
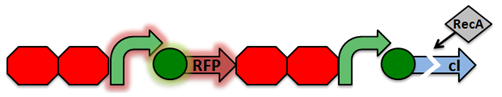
The design of our sensor was based on the lambda phage lysogenic vs lytic switch. Optimally, the system would be activated in the presence of DNA damage due to radiation. For that reason, we utilized the lambda phage switch, which transitions from the lysogenic cycle to the lytic cycle when DNA damage is detected. The genetic circuit we designed focuses on the DNA damage-sensitive lambda phage lytic-lysogenic cycle switch followed by a rapid response reporter similar to the immobilized fusion enzyme system pioneered by the Imperial College of London 2010 iGEM team. An initial design of our sensor circuit is illustrated below.

Under normal conditions, PRM is active, which transcribes the lambda repressor, cI + RBS. The lambda repressor binds to OR1 and OR2, repressing PR, which in turn inhibits the transcribtion of Cro and TEV.
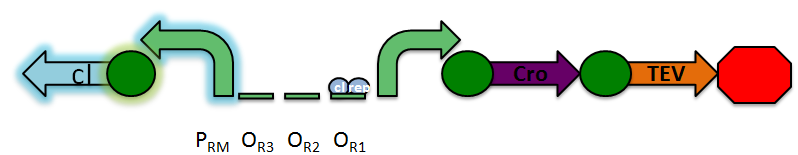
Once DNA damaged occurs, the ssDNA binds to RecA. RecA becomes activated and cleaves the cI repressor, allowing for PR to turn on. Cro then binds to OR3, which represses PRM and inhibits the transcription of the cI repressor. Now that PR is active, the transcription of the TEV protease occurs, which is used in the reporter structure of the circuit.
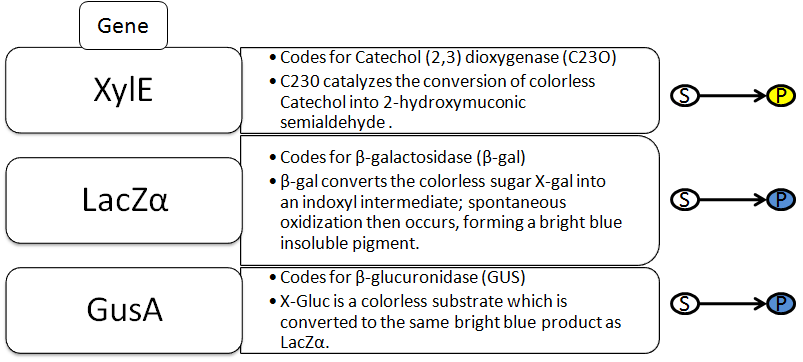
The Reporter
RecA Co-Protease
The normal laboratory strain of E. coli, DH10B, contains an inactivated form of RecA known as RecA1. Therefore, a series of mutations must occur in order to restore its proteatic function and reduce recombination.
Activation of RecA
- Point Mutation at b.p 720: A ⇒ G
Removal of Recombinase Activity
- Arginine 243 ⇒ Glutamine
- Lysine 286 ⇒ Asparagine
In order to use the standard bio-bricking techniques, we also had to remove these naturally occurring enzymatic restriction sites
- PST1 Site [CTGCAG]
- CAG ⇒ CTG (Codon usage bias decreases: .69 to .31)
- EcoR1 Site [GAATTC]
- TTC ⇒ TTT (Codon usage bias increases: .49 to .51)
Activated RecA then cleaves the cI repressor, causing transcription of RFP. However, if RecA exhibits recombinant activity, then the homologous double terminator regions will recombine. This causes the deletion of the repressible promoter and RFP.
Part 3
Results
| Home | Team | Penn State Official Team Profile | Project | Parts Submitted to the Registry | Modeling | Notebook | Safety | Attributions |
|---|
 "
"