Team:Cambridge/Experiments/Initial Exercise Group One
From 2011.igem.org
(→Reasons for choice) |
(→Construct Design) |
||
| Line 18: | Line 18: | ||
===Construct Design=== | ===Construct Design=== | ||
We wish to: | We wish to: | ||
| - | *keep the original promotor for | + | *keep the original promotor for DivIVA* |
| - | *remove GFP promotor sequence* | + | *remove the GFP promotor sequence* |
| - | *remove DivivA stop codon* | + | *remove the DivivA stop codon so that GFP is translated together with the DivIVA protein* |
| - | *Remove ComGA stabiliser sequence* | + | *Remove the ComGA stabiliser sequence pre-existing on the plasmid vector* |
| - | + | ‘Linker’ code was also used by Edwards and Errington - however, we suspect that this is related to the restriction enzyme assembly process they used and hence we didn’t include it in our design. | |
[[File:Cam_Oligo design_sketch.jpeg|frameless|700px|A sketch of design concept]] | [[File:Cam_Oligo design_sketch.jpeg|frameless|700px|A sketch of design concept]] | ||
Revision as of 16:45, 9 July 2011
Contents |
Initial Exercise: Katy, Heather, Marta and Veronica
The aim of the project was to produce interesting patterns of fluorescence in a colony of Bacillus Subtilis bacteria by fusion of a single gene of interest in the bacillus ge=nome to GFP using Gibson Assembly. These patterns would need to be clearly observable using a confocal microscope. After some research, our group decided to fuse GFP to the DivIVA gene.
Reasons for choice
We decided to perform this fusion for a number of reasons. Firstly, DivIVA plays and interesting role in cell division; it is believed to control septum positioning in Bacillus Subtilis. This was investigated (in [http://onlinelibrary.wiley.com/doi/10.1046/j.1365-2958.1997.3811764.x/abstract this] study by Edwards and Errington) by considering the phenotype of mutants produced when certain bases in the gene were altered.
Mutants showed:
- inhibition of cell division initiation*
- abnormally placed septum*
- some small anucleate cells*
We also hoped that this fusion would have a high chance of success, since a successful DivIVA-GFP fusion had already been documented, indicating that fluorescence was constitutively expressed at all sites of cell division, new and old, in the bacteria. We therefore know what characteristics to expect and so we should be able to determine whether or not the experiment was successful almost immediately under the microscope.
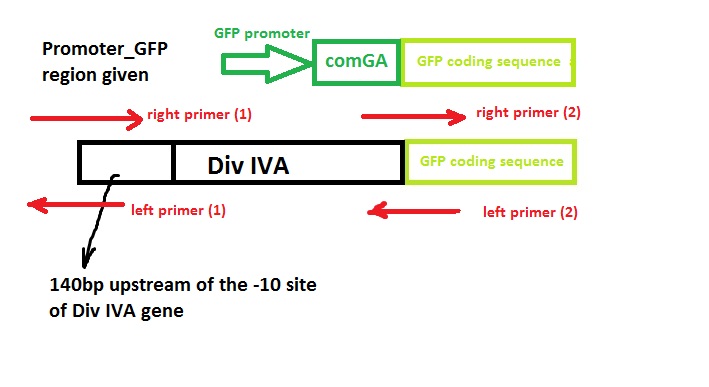
Construct Design
We wish to:
- keep the original promotor for DivIVA*
- remove the GFP promotor sequence*
- remove the DivivA stop codon so that GFP is translated together with the DivIVA protein*
- Remove the ComGA stabiliser sequence pre-existing on the plasmid vector*
‘Linker’ code was also used by Edwards and Errington - however, we suspect that this is related to the restriction enzyme assembly process they used and hence we didn’t include it in our design.
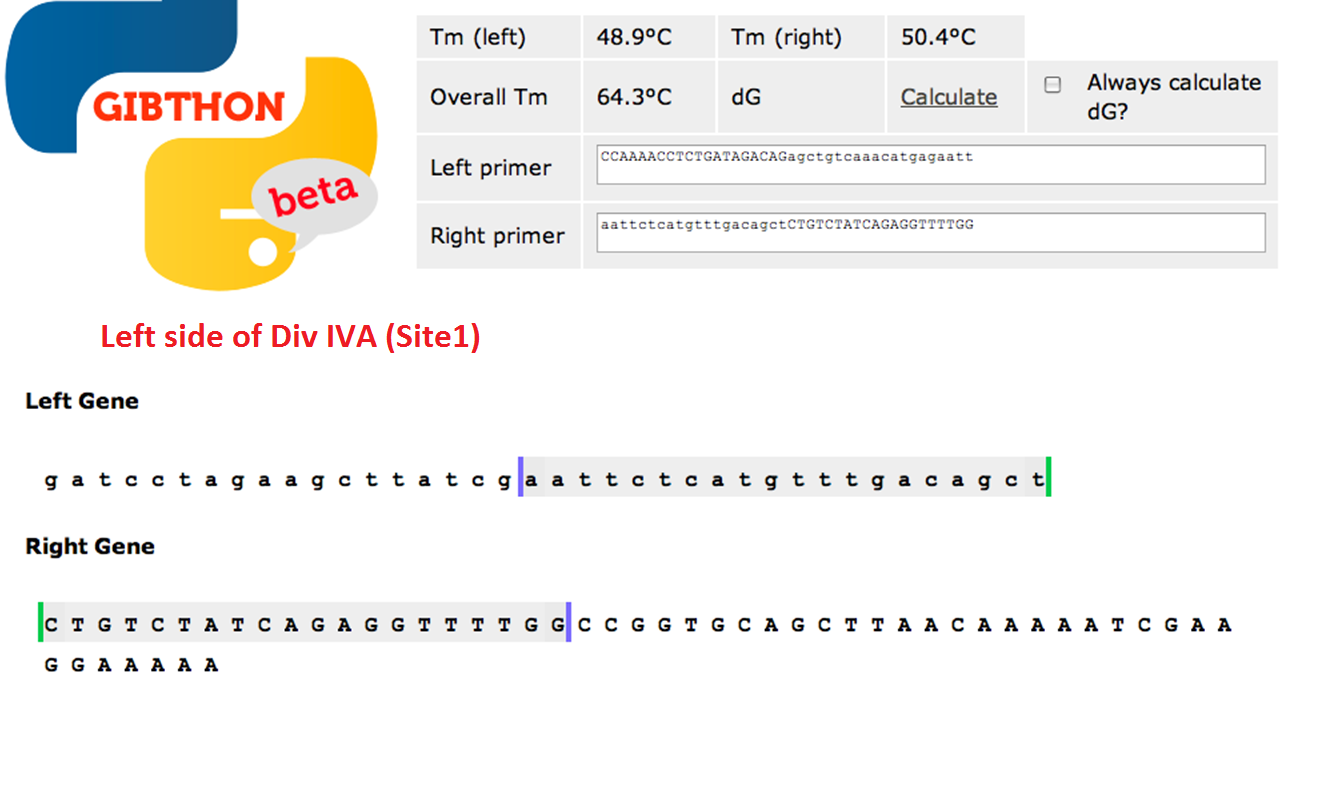
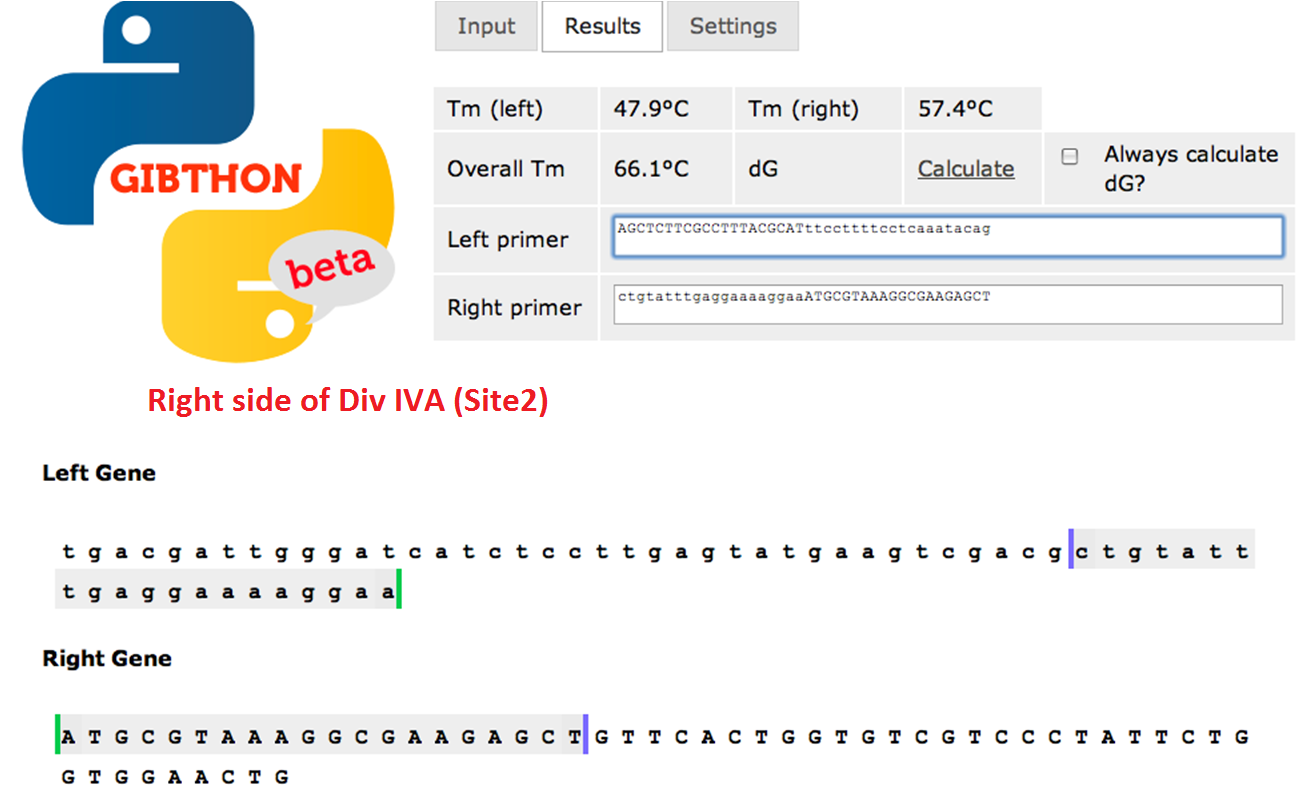
Primer Design
used the Gibthon Construct Designer developed by the 2010 iGEM Cambridge team
Left side of the Div IVA coding sequence(site 1)
Right side of the Div IVA coding sequence(site 2)
 "
"