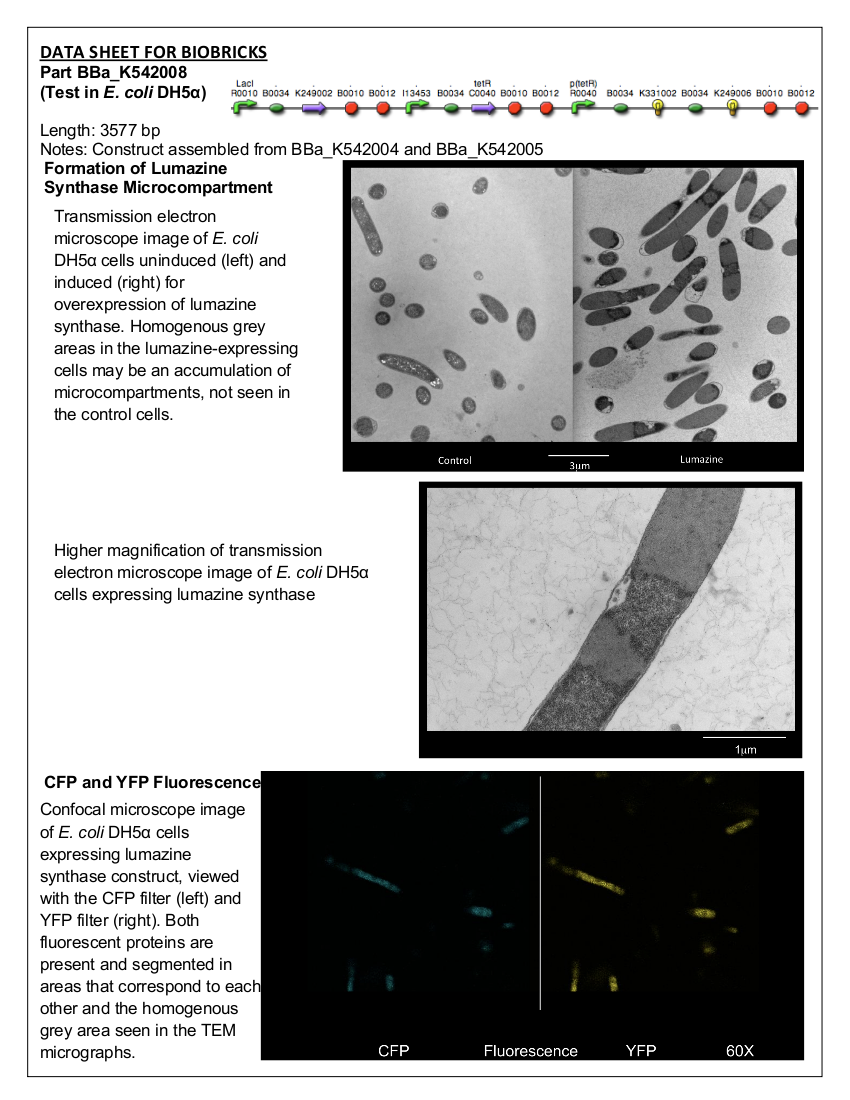
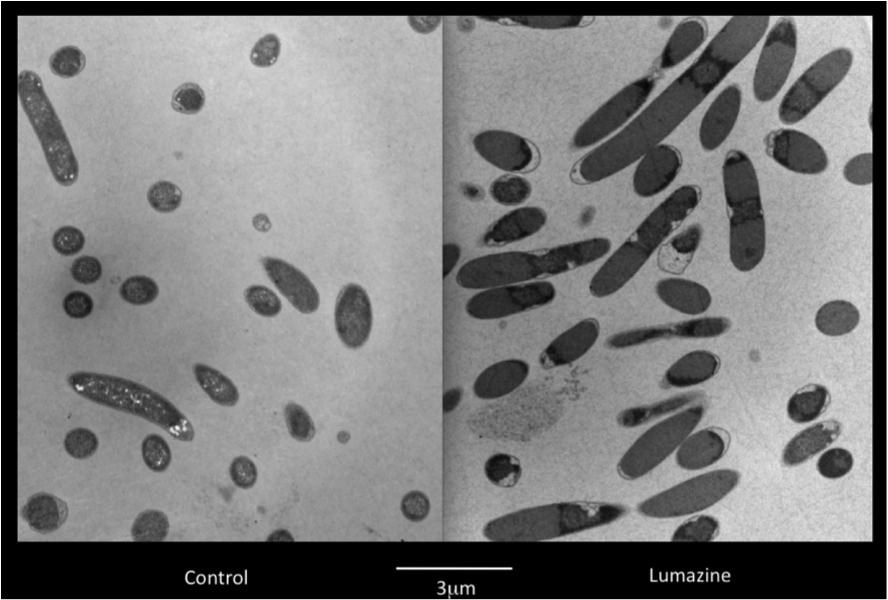
Team:Lethbridge/Results
From 2011.igem.org
Liszabruder (Talk | contribs) (→Part: BBa_K542009 – Antigen 43) |
|||
| (17 intermediate revisions not shown) | |||
| Line 91: | Line 91: | ||
[[image:uoflECFluorescence.png|center|400px]] | [[image:uoflECFluorescence.png|center|400px]] | ||
<br> | <br> | ||
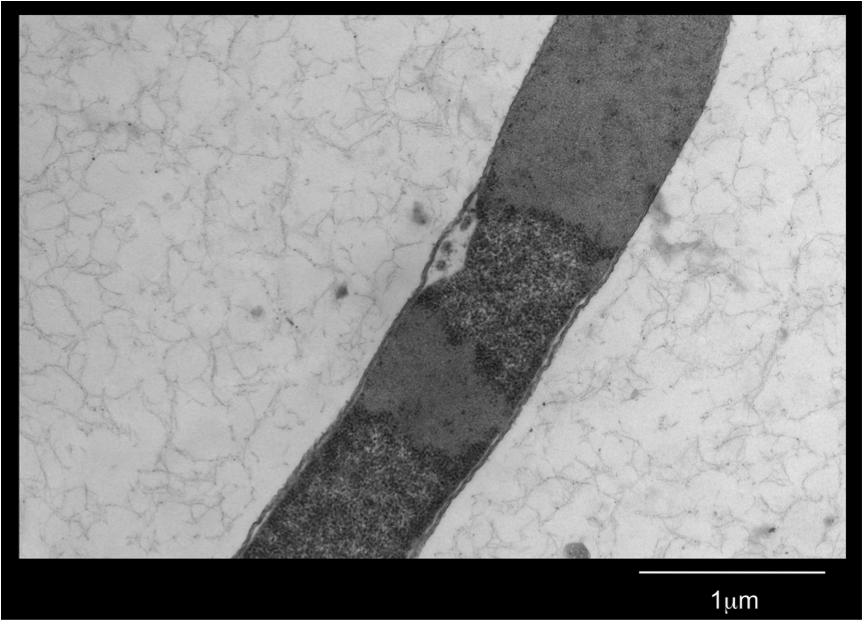
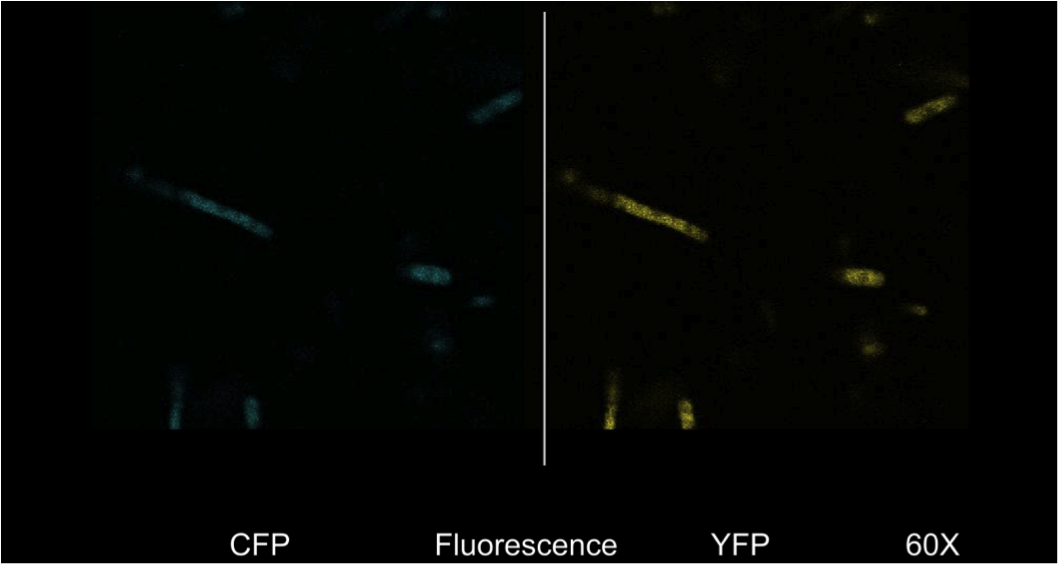
| - | <b>Figure 4.</b> <i>E. | + | <b>Figure 4.</b> <i>E. coli</i> DH5a cells containing the BBa_K542008 construct as viewed under spectral confocal microscope with 60X magnification. The left picture corresponds to the ECFP filter and the right picture corresponds to the EYFP filter. |
| + | |||
====Conclusion==== | ====Conclusion==== | ||
| - | Figure 4 shows that the fluorescent proteins were expressed in the cells. It is also evident that the fluorescence was segmented in a way that looks similar to that found in the homogenous grey areas in the TEM micrographs of the cells. | + | Figure 4 shows that the fluorescent proteins were expressed in the cells. It is also evident that the fluorescence was segmented in a way that looks similar to that found in the homogenous grey areas in the TEM micrographs of the cells. |
| + | |||
| + | ===Scanning Electron Microscopy=== | ||
| + | |||
| + | We attempted to view lumazine synthase microcompartments under scanning electron microscopy. However, because our microcompartments were in solution, the moisture remaining caused the electron beam to heat the sample. This destroyed the entire sample, including the carbon material it was placed on. | ||
| + | |||
| + | [[image:uoflsem.png|center]] | ||
| + | |||
| + | <b>Figure 1.</b> Scanning electron micrograph of lumazine synthase. The microcompartments are not visible due to the electron beam destroying the sample, as seen in the above dark spots. | ||
| + | |||
| + | ===Fluorescent Resonance Energy Transfer (FRET)=== | ||
| + | ====Materials and Methods==== | ||
| + | <i>Preparation of Cell Cultures and Controls</i><br> | ||
| + | A total of six 5ml cell cultures of Escherichia coli DH5α cells containing the BBa_K542008 construct were grown under different conditions overnight at 37°C. As controls parts BBa_K542006, BBa_K331031, BBa_K331033, BBa_K542001 were grown normally overnight at 37°C.<br> | ||
| + | |||
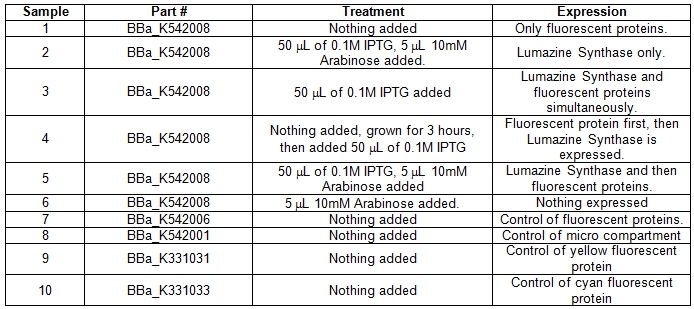
| + | <b>Table 1.</b> E.coli DH5α cells cultures grown in LB media and incubated overnight with shaking at 37°C. Samples #1-6 was grown up differently with specific treatments for the expression of certain parts within the BBa_K542008 construct. Samples #7-10 was grown as controls. <br> | ||
| + | [[image:UolFRETTable1.jpg|center|600px]]<br><br> | ||
| + | |||
| + | The following morning, six 50mL cultures were inoculated with 1mL of overnight culture grown to sample 6 (with nothing expressed), and four 50mL cultures were inoculated with samples 7-10. The 50mL cultures were allowed to grow with shaking at 37oC until they reached an OD600 of 0.6 (approximately 3 hours). Samples 1-3 and 6-10 were immediately pelleted by centrifugation (4oC, 5000xg, 20min) and placed on ice. Samples 4 and 5 were treated as described in table above. Pellets were weighed, and resuspended in 10mL/mg wet weight of the following buffer: | ||
| + | *50mM TrisHCl (pH8.0) | ||
| + | *250mM NaCl | ||
| + | *5mM 2ME | ||
| + | In order to evaluate expression levels, 1mL of cells was removed from each culture, and analysed using SDS-PAGE (12%).<br><br> | ||
| + | |||
| + | <i>Lysis of Cells and Clearing of Cell Lysate</i><br> | ||
| + | Resuspended cell cultures were incubated on ice, and subjected to 3x20 pulse sonication bursts, with 3 minute rests on ice.<br> | ||
| + | Lysate was cleared by centrifugation (4C, 30000xg, 1 hour). Cleared cell lysate was removed and retained for fluorescent analysis.<br><br> | ||
| + | <i>Scanning of Cell Lysate using a Spectrofluorometer</i><br> | ||
| + | Using a spectrofluorometer, 3 mL of cell lysate was used for each scan. Each scan was done at 5nm excitation and 5nm emission slits. For the analysis of the expression of Förster resonance energy transfer (FRET) within the micro compartment, each cell lysate was excited at 439nm and the emission spectra was read from 444nm to 650nm. | ||
| + | |||
| + | ====Results==== | ||
| + | <i>Expression levels of Lumazine Synthase and fluorescent proteins</i><br><br> | ||
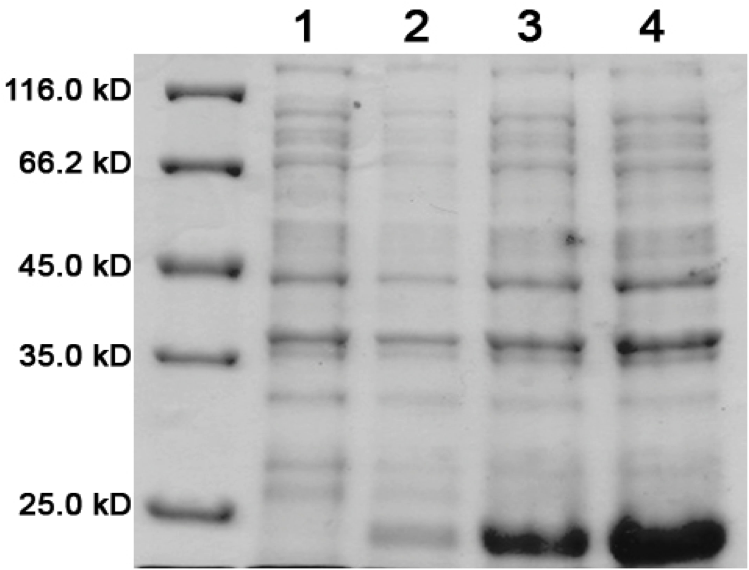
| + | Load order for the 12% denaturing polyacrylamide gel is as follows:<br> | ||
| + | # Low Range Molecular Marker | ||
| + | # K542008 – No Expression (Sample 6) | ||
| + | # K542001 – Lumazine Synthase Control (Sample 8) | ||
| + | # K331031 – YFP Control (Sample 9) | ||
| + | # K331033 – CFP Control (Sample 10) | ||
| + | # K542006 – Fluorescent Proteins (Both YFP and CFP) Control (Sample 7) | ||
| + | # K542008 – Expression of fluorescent proteins only (Sample 1) | ||
| + | # K542008 – Expression of Lumazine Synthase only (Sample 2) | ||
| + | # K542008 – Co-expression of Lumazine Synthase and fluorescent proteins (Sample 3) | ||
| + | # K542008 – Fluorescent proteins first, then Lumazine Synthase (Sample 4) | ||
| + | # K542008 – Lumazine Synthase first, then fluorescent proteins (Sample 5) | ||
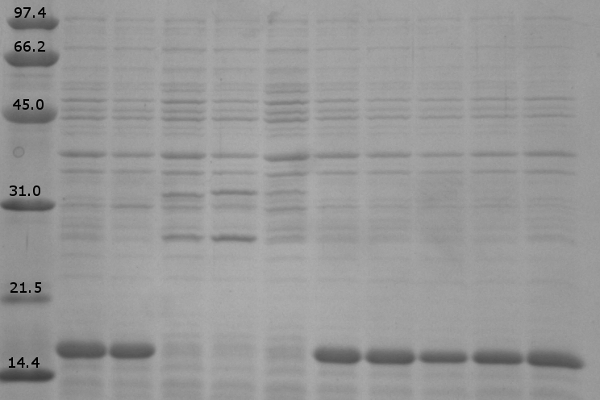
| + | [[image:UolK542008ExpressionPatterns.jpg|center|400px]] | ||
| + | <b>Figure 1.</b> Expression patterns of K542008 and controls<br><br> | ||
| + | In every K542008 sample, regardless of treatment, lumazine synthase (16.6kDa) is expressed. This means that we are unable to control production of lumazine synthase as we had hoped. By altering the promoter, we hope to be able to control the production of lumazine synthase in order to evaluate how the expression patterns of lumazine synthase and arginine tagged proteins affect efficiency of co-localization of tagged proteins within the compartment.<br><br> | ||
| + | The expression of the fluorescent proteins (each approximately 27kDa) can easily been seen in lanes 2 and 3. Although it is difficult to see fluorescent proteins in K542008 samples, fluorescence observed in FRET experiments suggests that protein is expressed.<br> <br><br> | ||
| + | |||
| + | <i>FRET</i><br><br> | ||
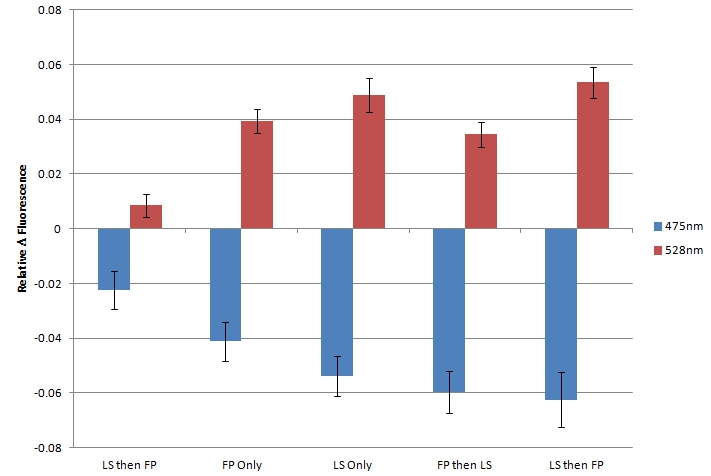
| + | By using sample 7 (fluorescent protein control; BBa_K542006) as the baseline for expression of fluorescent proteins in the cleared cell lysates, we evaluated samples 1-5 for FRET by comparing fluorescence at 475nm (CFP) and 528nm (YFP). <br> | ||
| + | [[image:UolK542008FRET.jpg|center|600px]] | ||
| + | <b>Figure 2.</b> Relative change in fluorescence at 475nm and 528nm, when excited by light at 439nm. "LS then FP" is Sample 5; "FP Only" is Sample 1; "LS Only" is Sample 2; "FP then LS" is Sample 3; "LS then FP" is Sample 4.<br><br> | ||
| + | |||
| + | In each sample, we observed a decrease in fluorescence at 475nm and a simultaneous increase in fluorescence at 528nm. We believe that this decrease in CFP fluorescence (observed at 475nm) and concomitant increase in YFP fluorescence (observed at 528nm) is indicative of FRET interactions occurring between CFP and YFP. The presence of FRET interactions naturally leads us to the conclusion that the oligo-arginine-tagged CFP and YFP are being co-localized within the lumazine synthase microcompartment. <br> | ||
| + | Within the FRET results, the condition where lumazine synthase was expressed before the fluorescent proteins were expressed shows lower levels of fluorescent change than the other conditions, suggesting that fluorescent proteins have difficulty entering the completely formed microcompartments. | ||
| + | |||
| + | ====Conclusion==== | ||
| + | |||
| + | # Proteins that have an oligo-arginine tail fused to their C-termini are localized within the microcompartment formed by the oligomerization of lumazine synthase proteins | ||
| + | # Proteins can localize within the compartment more easily when they are co-expressed with lumazine synthase | ||
| + | :::Proteins have a more difficult time entering the compartment when the compartment is fully formed | ||
| + | |||
| + | ====Discussion==== | ||
| + | While we hoped to be able to control the expression of the fluorescent proteins and lumazine synthase temporally, our SDS-PAGE gel indicates that even in the absence of IPTG, the <i>lac</i>-inducible lumazine synthase is being expressed. This means we are not able to achieve one of our conditions – fluorescent proteins expressed first, then lumazine synthase. The constant expression of lumazine synthase gave us several measurements with co-expression of lumazine synthase and fluorescent proteins. Furthermore, co-expression approximates the condition we were attempting for our first condition – namely, the formation of microcompartments around tagged proteins. We were also able to measure the condition where lumazine synthase was expressed in the absence of fluorescent proteins, which were expressed after the production of the lumazine synthase microcompartments. We interpret the results to indicating that tagged proteins have more difficulty entering pre-formed lumazine synthase microcompartments.<br><br> | ||
| + | |||
==References== | ==References== | ||
(1) Seebeck, F., Woycechowsky, K., Zhuang, W., Rabe, J., and Hilvert, D. (2006). A simple tagging system for protein encapsulation. Journal of the American Chemical Society. 128: 4516-4517. | (1) Seebeck, F., Woycechowsky, K., Zhuang, W., Rabe, J., and Hilvert, D. (2006). A simple tagging system for protein encapsulation. Journal of the American Chemical Society. 128: 4516-4517. | ||
| Line 152: | Line 218: | ||
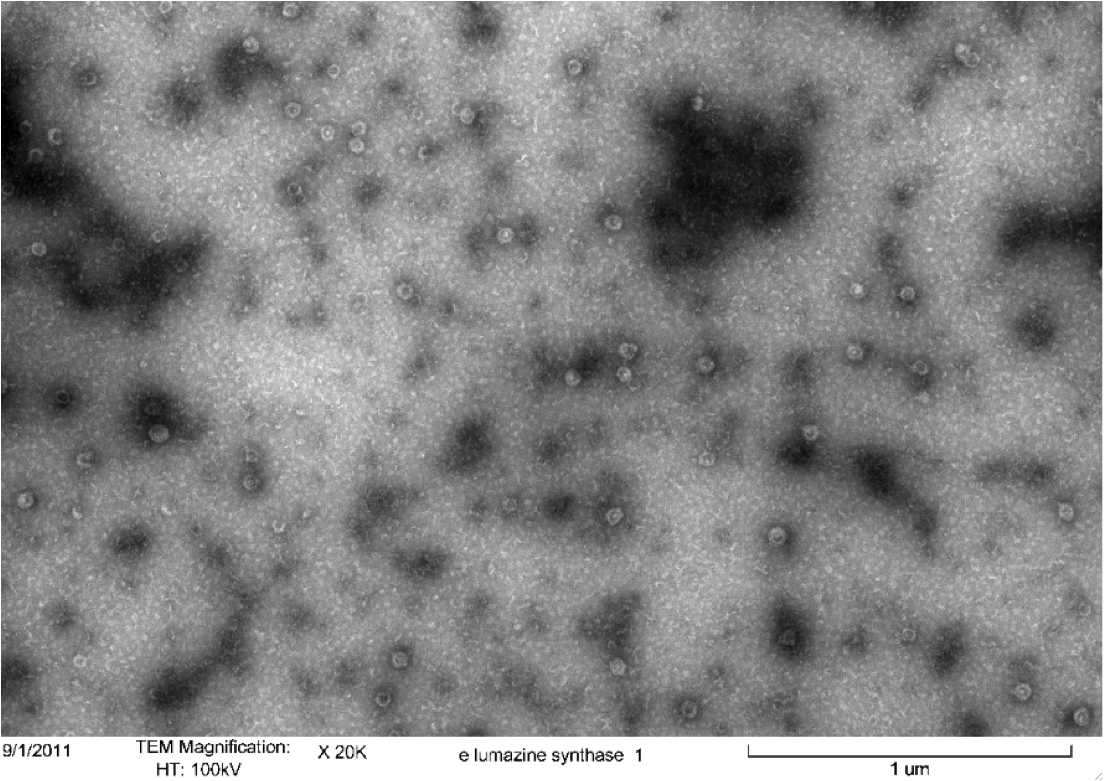
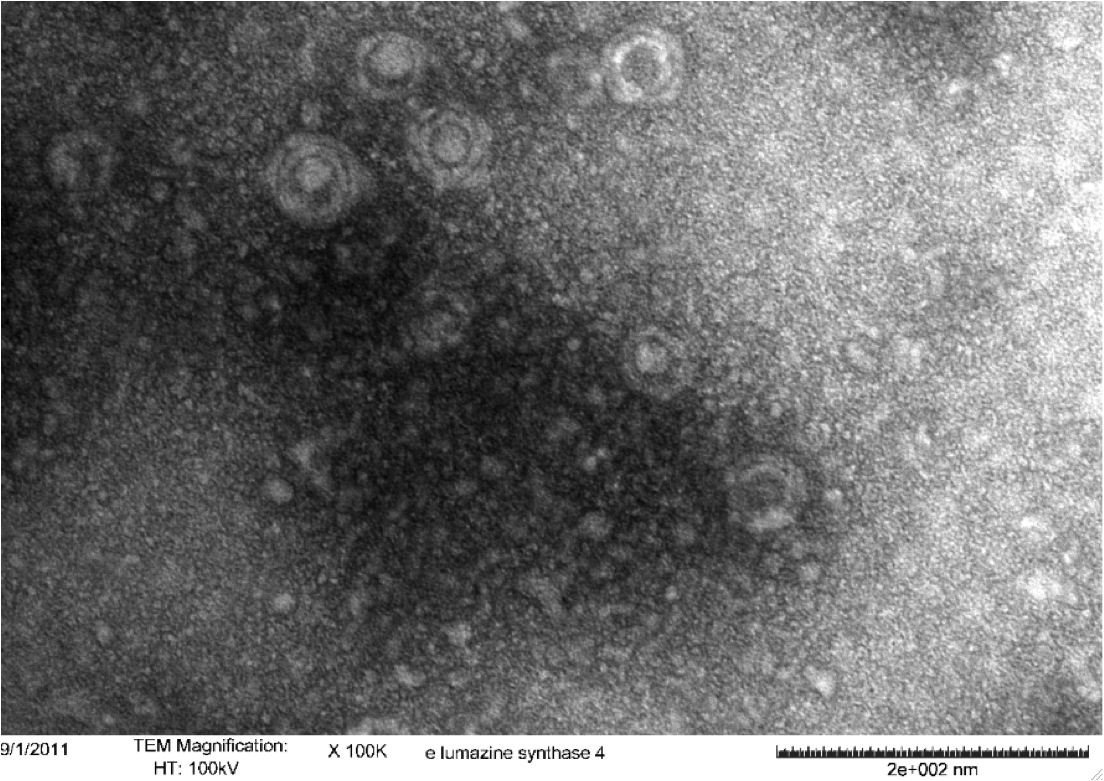
<b>Figure 5.</b> Transmission electron microscopy of Enhanced Lumazine Synthase microcompartments. Microcompartments of approximately 40 nm can be seen at x 100K magnification. | <b>Figure 5.</b> Transmission electron microscopy of Enhanced Lumazine Synthase microcompartments. Microcompartments of approximately 40 nm can be seen at x 100K magnification. | ||
====Conclusion==== | ====Conclusion==== | ||
| - | As expected, TEM micrographs show polyhedral particles of approximately 40 nm, corresponding to the size described by Wörsdörfer et al. (2011). This confirms that E. coli is capable of producing enhanced lumazine synthase. | + | As expected, TEM micrographs show polyhedral particles of approximately 40 nm, corresponding to the size described by Wörsdörfer et al. (2011). This confirms that ''E. coli'' is capable of producing enhanced lumazine synthase. |
| + | |||
==References== | ==References== | ||
| Line 193: | Line 260: | ||
[[image:uoflBamHIdatasheet.png|100px|center]] | [[image:uoflBamHIdatasheet.png|100px|center]] | ||
==Introduction== | ==Introduction== | ||
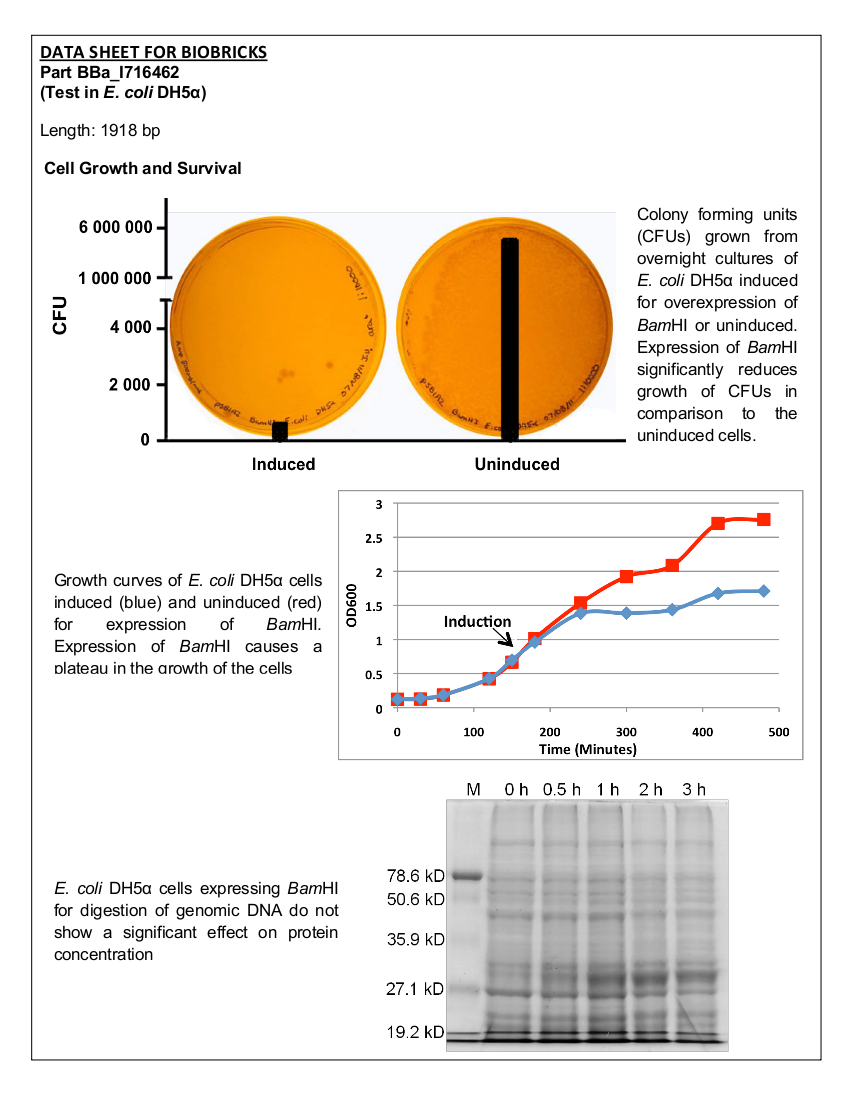
| - | Part I716462 was previously characterized by the UC Berkeley iGEM team in 2007. The purpose of this part was to prevent unwanted cell proliferation by engineering a genetic self-destruct mechanism into bacteria that upon induction would express a genetic material-degrading toxin, the endonuclease BamHI that would kill cells yet preserve their physical integrity. | + | Part I716462 was previously characterized by the UC Berkeley iGEM team in 2007. The purpose of this part was to prevent unwanted cell proliferation by engineering a genetic self-destruct mechanism into bacteria that upon induction would express a genetic material-degrading toxin, the endonuclease BamHI that would kill cells yet preserve their physical integrity. <i>Bam</i>HI has a restriction cut site of G'GATCC, generating sticky ends (1). The restriction endonuclease was placed under the control of the arabinose- inducible promoter, pBAD. In 2007, the UC Berkeley iGEM team was able to show that cells lost their ability to reproduce but did not lyse upon induction with arabinose (2). The University of Lethbridge 2011 iGEM team wished to further characterize this part to degrade the genomic DNA of E. coli in order to have a functional chassis for our tailings pond clean-up kit without the risk of DNA contamination. In this way, tight control and regulation over our engineered bacterial cells is possible. |
| + | <br><br> | ||
| + | Registry Entry - BBa_I716462 | ||
| + | <br><br> | ||
| + | Length – 1918 bp | ||
==Characterization== | ==Characterization== | ||
===Cell Survival=== | ===Cell Survival=== | ||
| Line 201: | Line 272: | ||
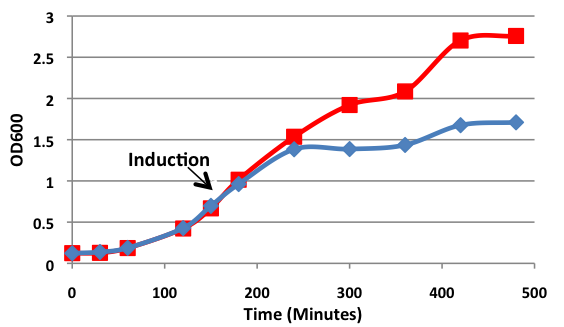
[[image:uoflBamHIgrowthcurve.png|center|400px]] | [[image:uoflBamHIgrowthcurve.png|center|400px]] | ||
<br><br> | <br><br> | ||
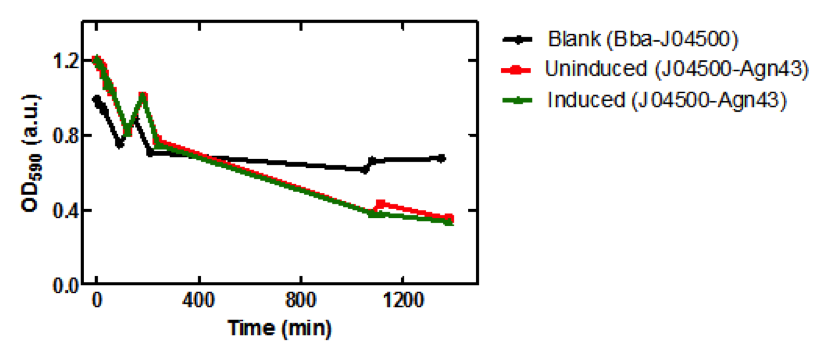
| - | <b>Figure 1.</b> | + | <b>Figure 1.</b> Growth curves of <i>E. coli</i> DH5α cells induced (blue) and uninduced (red) for expression of <i>Bam</i>HI. Cultures were induced at an OD<sub>600</sub> of 0.60. |
<br><br> | <br><br> | ||
[[image:uoflBamHICFU.png|center|400px]] | [[image:uoflBamHICFU.png|center|400px]] | ||
<br><br> | <br><br> | ||
| - | <b>Figure 2. </b><i>E.coli</i>DH5α cells containing the <i>Bam</i>HI construct from overnight cultures. The samples were plated on LB agar plates and the number of colony forming units (CFU) was counted. Left: culture induced for | + | <b>Figure 2. </b><i>E. coli</i> DH5α cells containing the <i>Bam</i>HI construct from overnight cultures. The samples were plated on LB agar plates and the number of colony forming units (CFU) was counted. Left: culture induced for <i>Bam</i>HI expression with arabinose, right: uninduced culture. |
===Analysis of Genomic DNA=== | ===Analysis of Genomic DNA=== | ||
====Materials and Methods==== | ====Materials and Methods==== | ||
| - | <i>E. coli</i> DH5α cells were grown and induced to overexpress one of three samples: <i>Bam</i>HI construct induced with arabinose ( | + | <i>E. coli</i> DH5α cells were grown and induced to overexpress one of three samples: <i>Bam</i>HI construct induced with arabinose (10µM) at an OD<sub>600</sub> of 0.6, <i>Bam</i>HI construct uninduced, non-<i>Bam</i>HI construct induced with arabinose (10µM) at OD<sub>600</sub> of 0.6. Cells were grown in LB media with 100 ng/mL ampicillin. |
<br><br> | <br><br> | ||
| - | Qiagen Blood and Tissue Kit was used to isolate the genomic DNA of gram-negative bacteria. A 0.8% agarose gel was ran at 150 V for 90 minutes to separate the genomic DNA of <i>E. coli</i> DH5α cells collected at different time points (pre-induced and 30 min, 1 hr, 2 hr, and 3 hr post induction) for all three samples. | + | Qiagen Blood and Tissue Kit was used to isolate the genomic DNA of gram-negative bacteria. A 0.8% agarose gel was ran at 150 V for 90 minutes to separate the genomic DNA of <i>E. coli</i> DH5α cells collected at different time points (pre-induced and 30 min, 1 hr, 2 hr, and 3 hr post induction) for all three samples. One absorbance unit of cells were collected and analyzed by SDS-PAGE to monitor cellular protein levels. |
| + | |||
====Results==== | ====Results==== | ||
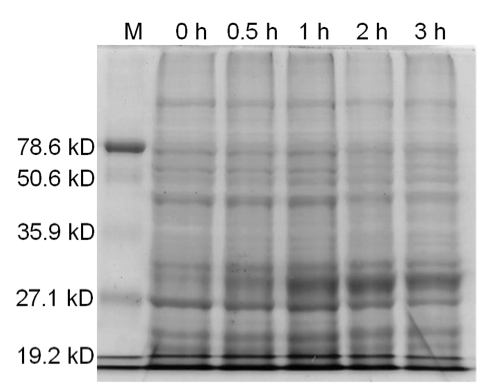
[[image:uoflBamHIgenomicDNA.png|center|400px]] | [[image:uoflBamHIgenomicDNA.png|center|400px]] | ||
<br> | <br> | ||
| - | <b>Figure 3.</b> | + | <b>Figure 3.</b> Induced <i>Bam</i>HI. 12% SDS-PAGE of cell lysate from <i>E. coli</i> DH5α cells containing BBa_I716462 induced for overexpression of <i>Bam</i>HI. Left to right: protein marker, 0 h, 0.5 h, 1 h, 2 h, and 3 h after induction with lactose. |
| - | + | ||
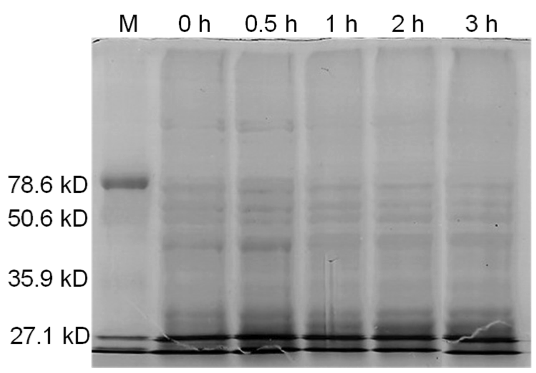
[[image:uoflBamHIfigure4.png|center|400px]] | [[image:uoflBamHIfigure4.png|center|400px]] | ||
<br> | <br> | ||
| - | <b>Figure 4.</b> Induced <i>Bam</i>HI. 12% SDS-PAGE of cell lysate from <i>E. coli</i> DH5α cells containing BBa_I716462 | + | <b>Figure 4.</b> Induced <i>Bam</i>HI. 12% SDS-PAGE of cell lysate from uninduced <i>E. coli</i> DH5α cells containing BBa_I716462. Left to right: protein marker, 0 h, 0.5 h, 1 h, 2 h, and 3 h after induction with lactose. |
<br><br> | <br><br> | ||
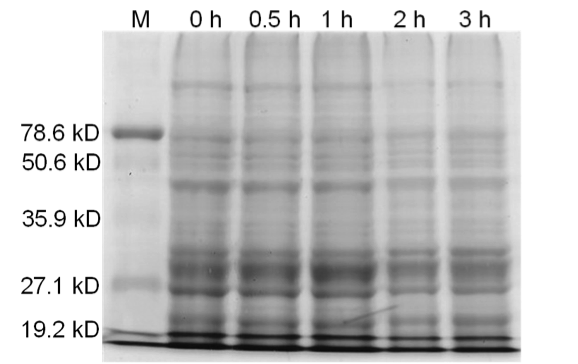
[[image:uoflBamHIfigure5.png|center|400px]] | [[image:uoflBamHIfigure5.png|center|400px]] | ||
<br> | <br> | ||
| - | <b>Figure 5.</b> Uninduced <i>Bam</i>HI. 12% SDS-PAGE of cell lysate from <i>E. coli</i> DH5α cells | + | <b>Figure 5.</b> Uninduced <i>Bam</i>HI. 12% SDS-PAGE of cell lysate from <i>E. coli</i> DH5α cells. Left to right: protein marker, 0 h, 0.5 h, 1 h, 2 h, and 3 h after induction with lactose. |
<br><br> | <br><br> | ||
| - | + | ==Conclusion== | |
| - | < | + | Upon induction with arabinose, <i>E. coli</i> DH5α cells containing the <i>Bam</i>HI construct did not reproduce as the overexpression of the <i>Bam</i>HI endonuclease digested the genomic DNA of the host cell. This is why in Figure 2 there are only 4 colonies seen as opposed to the control which shows a lawn of growth. In Figure 1, the optical density of the <i>Bam</i>HI expressing cells reaches a plateau; however, the same sample which has not been induced kept increasing, which indicates that the expression of <i>Bam</i>HI is affecting normal growth of the cells. |
| - | < | + | |
<br><br> | <br><br> | ||
| - | + | Samples analyzed by SDS-PAGE after induction of <i>Bam</i>HI expression show that protein concentration is not significantly affected (Figures 3-5). | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
==References== | ==References== | ||
(1) Viadiu H, Aggarwal AK (2000). Structure of BamHI bound to nonspecific DNA: a model for DNA sliding. Mol. Cell 5 (5): 889-895. | (1) Viadiu H, Aggarwal AK (2000). Structure of BamHI bound to nonspecific DNA: a model for DNA sliding. Mol. Cell 5 (5): 889-895. | ||
<br> | <br> | ||
| - | (2) iGEM 2007. (2011, May 10). BerkiGEM2007Present5. Retrieved from | + | (2) iGEM 2007. (2011, May 10). BerkiGEM2007Present5. Retrieved from https://2007.igem.org/BerkiGEM2007Present5. |
<br><br><br> | <br><br><br> | ||
| Line 245: | Line 311: | ||
==Results== | ==Results== | ||
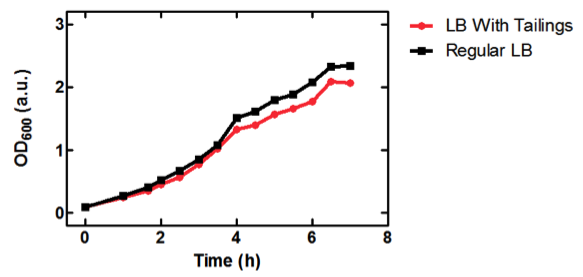
[[image:uofltailings.png|center|400px]] | [[image:uofltailings.png|center|400px]] | ||
| - | <b>Figure 1.</b> OD<sub>600</sub> of <i>E. coli</i> DH5α cells containing BBa_K331009 cultured in LB medium made with tailings water (red) and LB medium made with MilliQ H<sub>2</sub>O (black). | + | <b>Figure 1.</b> OD<sub>600</sub> of <i>E. coli</i> DH5α cells containing BBa_K331009 cultured in LB medium made with tailings water (red) and LB medium made with MilliQ H<sub>2</sub>O (black). Performed by our team. |
| + | <br><br> | ||
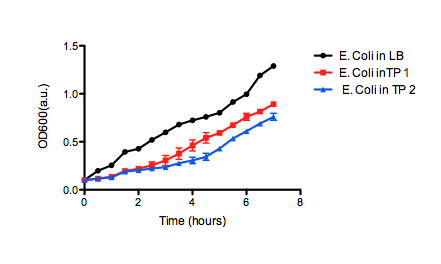
| + | [[image:uofltailings2.png|center|400px]] | ||
| + | <b>Figure 2.</b> OD<sub>600</sub> of <i>E. coli</i> DH5α cells containing BBa_K331009 cultured in LB medium made with tailings water (red and blue) and LB medium made with MilliQ H<sub>2</sub>O (black). Performed independently by the 2011 Calgary iGEM Team. | ||
| + | |||
==Conclusion== | ==Conclusion== | ||
As seen in Figure 1, <i>E. coli</i> DH5α cells were able to grow in LB medium made with tailings water, suggesting that the chassis will survive for use in tailings pond water. | As seen in Figure 1, <i>E. coli</i> DH5α cells were able to grow in LB medium made with tailings water, suggesting that the chassis will survive for use in tailings pond water. | ||
<br> | <br> | ||
<br> | <br> | ||
Latest revision as of 17:06, 12 April 2012
|
|
|
|---|
 "
"