Team:SJTU-BioX-Shanghai/Project/Subproject1-3
From 2011.igem.org
(Difference between revisions)
ChobitParrot (Talk | contribs) (→Note: Our device can be used as a regulating tool) |
(→Number of Rare Codons) |
||
| (4 intermediate revisions not shown) | |||
| Line 9: | Line 9: | ||
__NOTOC__ | __NOTOC__ | ||
| - | ==Number of Rare Codons== | + | ==Part I: Number of Rare Codons== |
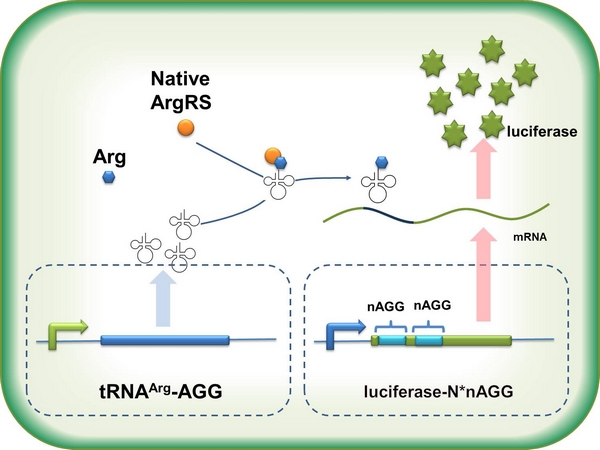
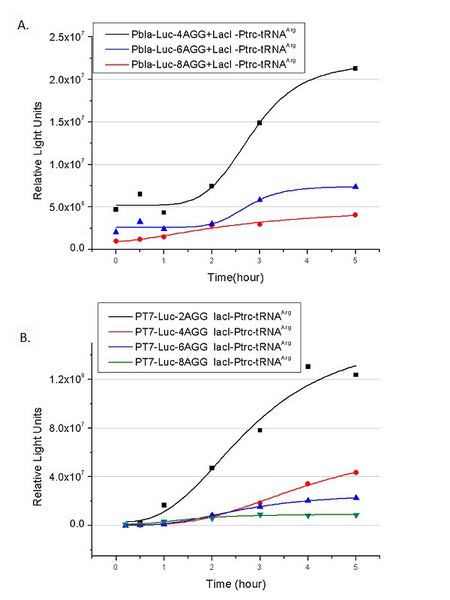
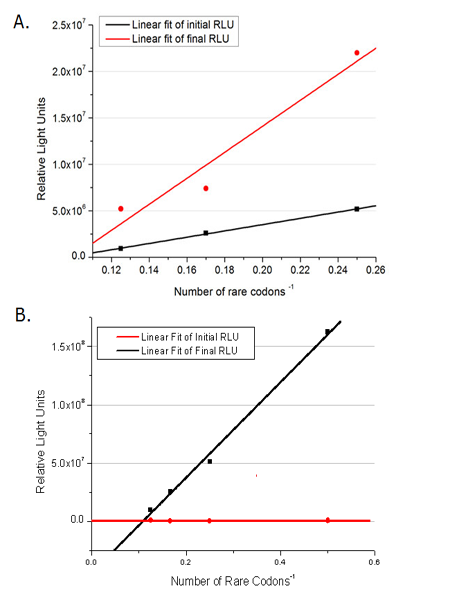
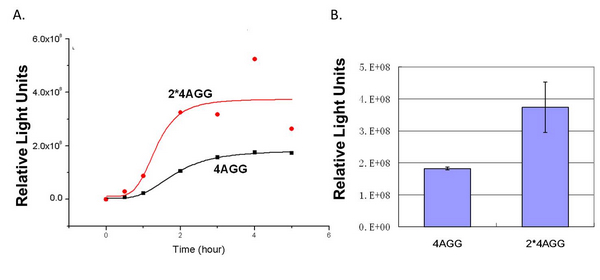
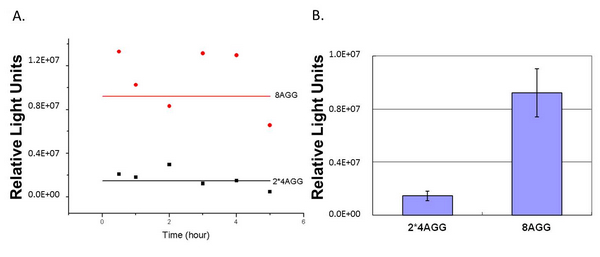
In this part we want to explore the influence of the number of rare codons inserted in the mRNA. We have inserted 2, 4, 6, 8 AGG codons respectively after the start codon in luciferase gene. T7 promoter or ''bla'' promoter<sup>[1]</sup> are used to control target protein mRNA amount. We use different combinations of number of AGG codons and strength of promoters to characterize regulation<sup>[1]</sup>. | In this part we want to explore the influence of the number of rare codons inserted in the mRNA. We have inserted 2, 4, 6, 8 AGG codons respectively after the start codon in luciferase gene. T7 promoter or ''bla'' promoter<sup>[1]</sup> are used to control target protein mRNA amount. We use different combinations of number of AGG codons and strength of promoters to characterize regulation<sup>[1]</sup>. | ||
| Line 63: | Line 63: | ||
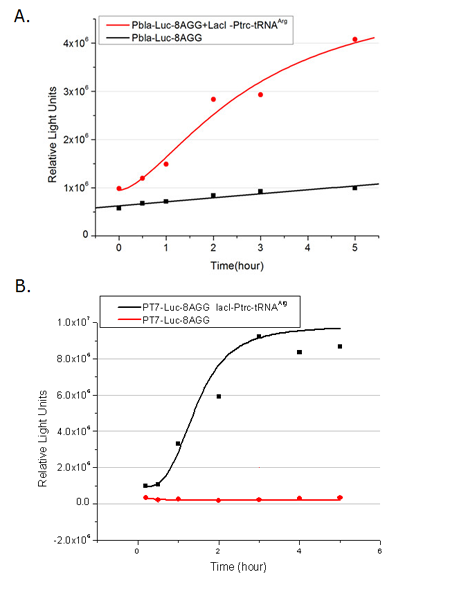
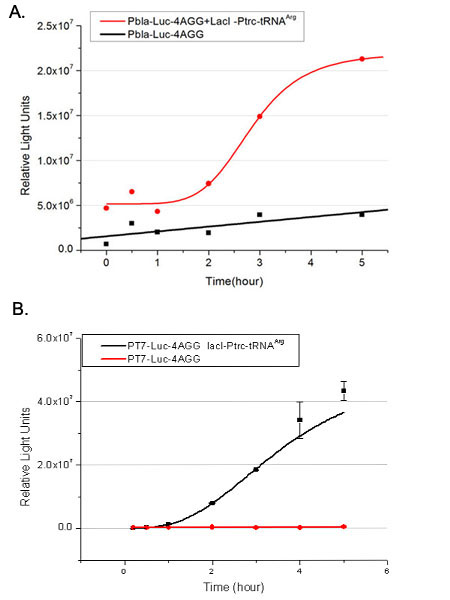
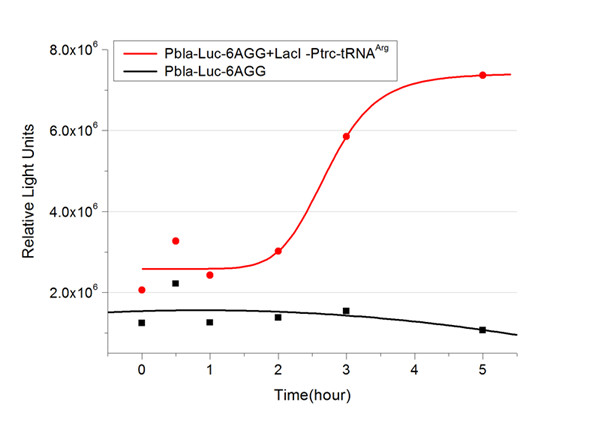
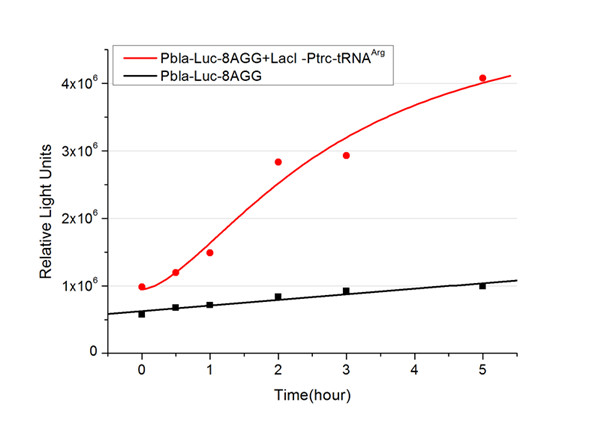
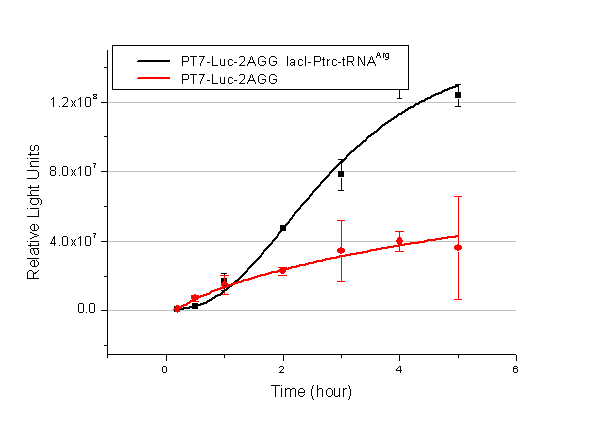
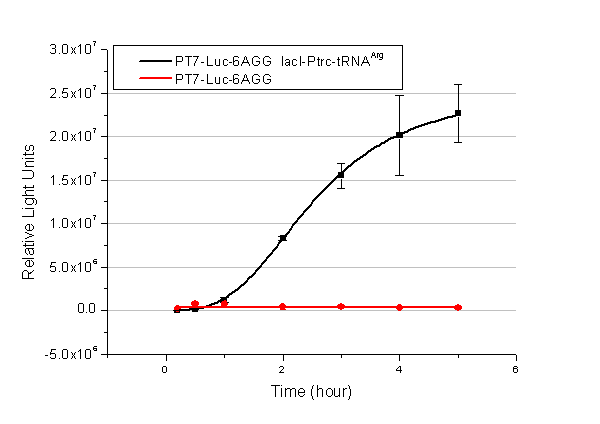
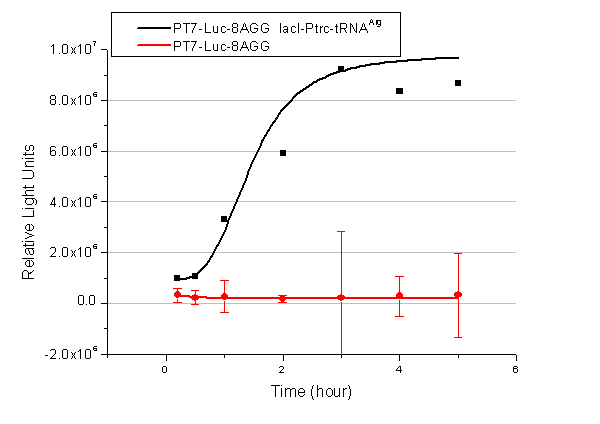
[[image:11sjtu_Compare_8.png|frame|center|''Fig.3'' (A) Luciferase (P''bla''-Luc-8AGG) production in cells overexpressing rare tRNA with ''lacI''-Ptrc-tRNA<sup>Arg</sup>. Luciferase production is reflected by bioluminescence emitted from the luciferin reaction. (B) Luciferase production in cells with PT7-Luc-8AGG as reporter. Here we analyze the influences of strong/weak promoter in luciferase production. Strong promoter (T7) of target gene can improve the titration curve, indicating that our device works better under strong target protein promoters. ]] | [[image:11sjtu_Compare_8.png|frame|center|''Fig.3'' (A) Luciferase (P''bla''-Luc-8AGG) production in cells overexpressing rare tRNA with ''lacI''-Ptrc-tRNA<sup>Arg</sup>. Luciferase production is reflected by bioluminescence emitted from the luciferin reaction. (B) Luciferase production in cells with PT7-Luc-8AGG as reporter. Here we analyze the influences of strong/weak promoter in luciferase production. Strong promoter (T7) of target gene can improve the titration curve, indicating that our device works better under strong target protein promoters. ]] | ||
| - | |||
==Location of Rare Codons== | ==Location of Rare Codons== | ||
| Line 81: | Line 80: | ||
[[image:11SJTU-3X4AGG.png|600px]] | [[image:11SJTU-3X4AGG.png|600px]] | ||
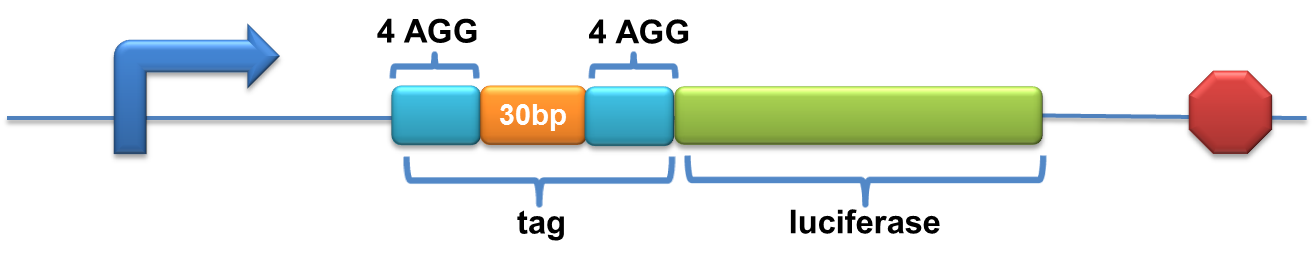
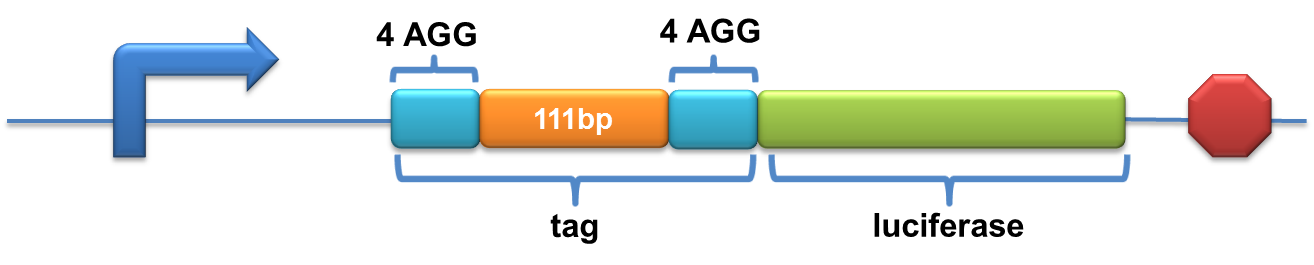
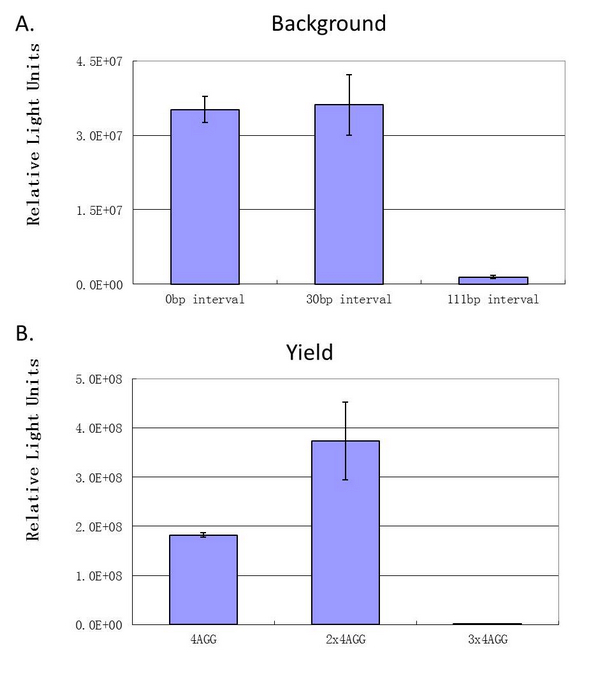
| - | Experiment results showed that | + | Experiment results showed that when there is 111bp interval between the two-copy 4AGG, background is lower than there is a 30bp interval. In another analysis, protein production can be induced to a higher level when there are two copies of AGG tandems. |
| - | + | ||
| - | + | ||
| - | [[image:11SJTU_rare_27.jpg|frame|center|''Fig.4'']] | + | [[image:11SJTU_rare_27.jpg|frame|center|''Fig.4 A. When there is 111bp interval between the two-copy 4AGG, background is lower than there is a 30bp interval. B. Protein production can be induced to a higher level when there are two copies of AGG tandems.'']] |
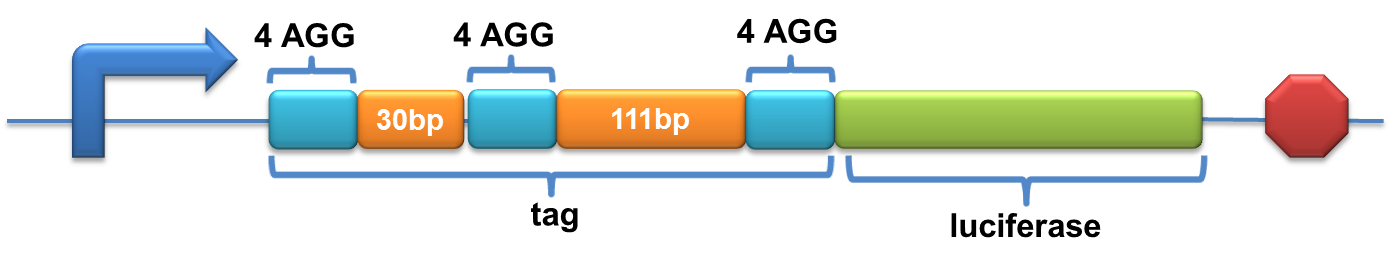
'''Experiment results showed that luciferase tagged with two-copy 4AGG insertions with an interval of 111 bp can increase yield and lower background noise of target protein.''' We further conducted experiments to test its characters. | '''Experiment results showed that luciferase tagged with two-copy 4AGG insertions with an interval of 111 bp can increase yield and lower background noise of target protein.''' We further conducted experiments to test its characters. | ||
| Line 122: | Line 119: | ||
From this experiment, we noticed that the typical working curve of our device can be better observed under IPTG induced ''lacI''-Ptrc-tRNA<sup>Arg</sup> ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K567001 BBa_K567001]) compared with UV excitation induced sulA promoter-tRNA<sup>Arg</sup>([http://partsregistry.org/wiki/index.php?title=Part:BBa_K567002 BBa_K567002]), though sulA promoter-tRNA<sup>Arg</sup> responded quicker to signals. So in the above experiments, we test with ''lacI''-Ptrc-tRNA<sup>Arg</sup>. | From this experiment, we noticed that the typical working curve of our device can be better observed under IPTG induced ''lacI''-Ptrc-tRNA<sup>Arg</sup> ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K567001 BBa_K567001]) compared with UV excitation induced sulA promoter-tRNA<sup>Arg</sup>([http://partsregistry.org/wiki/index.php?title=Part:BBa_K567002 BBa_K567002]), though sulA promoter-tRNA<sup>Arg</sup> responded quicker to signals. So in the above experiments, we test with ''lacI''-Ptrc-tRNA<sup>Arg</sup>. | ||
| + | |||
| + | ===Reference=== | ||
| + | [1]Ulrich Deuschlel., et al., Promoters of Escherichia coli: a hierarchy of in vivo strength indicates alternate structures The EMBO Journal vol.5 no. 11 pp.2987-2994, 1986 | ||
Latest revision as of 04:03, 29 October 2011
 "
"