User:Lytao/2
From 2011.igem.org
(→Artificial Innate Immunity System) |
|||
| (25 intermediate revisions not shown) | |||
| Line 2: | Line 2: | ||
{{Team:USTC-China/temp2}} | {{Team:USTC-China/temp2}} | ||
| + | == '''Artificial Innate Immunity System(AIIS)''' == | ||
| - | + | ||
| - | <html ><a href= "# | + | <html><a href= "#Background"> <u>1.Background</u> </a> </html><br/> |
| - | <html><a href= "# | + | <html><a href= "#Purpose"> <u>2.Purpose</u> </a> </html><br/> |
| - | <html><a href= "# | + | <html><a href= "#Principle"> <u>3.Principle</u> </a> </html><br/> |
| - | <html><a href= "# | + | <html><a href= "#Theoretical Design"> <u>4.Theoretical Design</u> </a> </html><br/> |
| - | <html><a name=" | + | <html><a href= "#Expermental Design and Results"> <u>5.Expermental Design and Results</u> </a> </html><br/> |
| - | == | + | <html><a href= "#Conclusion and Discussion"> <u>6.Conclusion and Discussion</u> </a> </html><br/> |
| + | |||
| + | |||
| + | |||
| + | <html><a name="Background" id=""></a></html> | ||
| + | |||
| + | ==Background== | ||
| + | <p> From mid May to June 2011, enteropathogenic E.coli brings a big panic to many different European countries. Why such an ordinary bacteria as E.coli can lead to a clinical catastrophe and kill a lot of people? The answer may be complicate, but the most important reason is that we can not find out the pathogens rapidly and can not apply the effective treatments.To solve this problem, we try to use the normal E.coli, which keep the symbiotic relationship with us humanbeings, to be a safety weapon to defend ourselves against pathogens.</p> | ||
<html> | <html> | ||
| - | <div class="center"><div class="thumb tnone"><div class="thumbinner" ><img alt=" | + | <div class="center"><div class="thumb tnone" style="width:352px;"><div class="thumbinner" style="width:352px;"><a href="/File:At().jpg" class="image"><img alt="At().jpg" src="/wiki/images/thumb/f/f0/At%28%29.jpg/350px-At%28%29.jpg" width="350" height="216" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:At().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure1.Enteropathogenic ''Escherichia coli''(left) brings us a big panic(right,Frank Bellew, New York Daily Graphic, September 29, 1873)</div></div></div></div> |
</html> | </html> | ||
| + | <p align=center class="ppp"></p> | ||
| + | <html><a name="Purpose" id=""></a></html> | ||
| + | ==Purpose== | ||
| + | <p> Using reprogrammed intestinal bacteria as a safety weapon to destroy the pathogens which invade the intestinal mucosal system.</p> | ||
| + | <html><a name="Principle" id=""></a></html> | ||
| + | ==Principle== | ||
| + | <p> As shown in Figure2, When pathogens invade the intestinal epithelium ,they will release some kind of chemical signal. After the reprogrammed intestinal bacteria capture this signal, such signal can activate the expression of the function gene ,then it will drive the reprogrammed intestinal bacteria to move toward the infection site from the original colony. Then the moving reprogrammed bacteria will express some lethal proteins at the infection site to kill pathogens by self-destruction, and the original colony will remain.</p> | ||
| + | <html><a name="Theoretical Design" id=""></a></html> | ||
| + | <html> | ||
| + | <head> | ||
| - | < | + | </head> |
| - | < | + | <div align="center"> |
| - | < | + | <object type="application/x-shockwave-flash" height=300 width=400 data="https://static.igem.org/mediawiki/2011/4/43/Application.swf" > |
| + | <param name="movie" value="https://static.igem.org/mediawiki/2011/4/43/Application.swf" /> | ||
| + | <param name="quality" value="high" /> | ||
| + | <param name="" value="Exactfit"/> | ||
| + | </object> | ||
| + | </div> | ||
| + | </html> | ||
| + | |||
| + | ==Theoretical Design== | ||
| - | + | <p>'''1.AI(Auto Inducer)as a Chemical Signal'''</p> | |
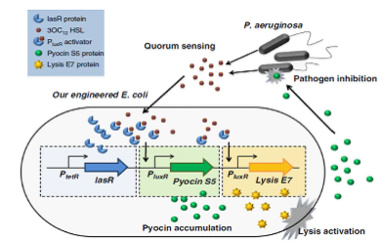
| + | <p> By using the device from the article ''Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen'' (Figure3.), we can modify our original design to achieve the function which we have described before.(As shown in Figure4.)</P> | ||
<html> | <html> | ||
| - | <div class="center" ><div class="thumb tnone" style="width: | + | <div class="center"><div class="thumb tnone" style="width:382px;"><div class="thumbinner" style="width:382px;"><a href="/File:V().jpg" class="image"><img alt="V().jpg" src="/wiki/images/thumb/a/a6/V%28%29.jpg/380px-V%28%29.jpg" width="380" height="242" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:V().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure3.This image comes from Nazanin Saeidi.etc (2011).</div></div></div></div> |
</html> | </html> | ||
| - | < | + | <p align=center class="ppp"></p> |
| - | + | ||
<html> | <html> | ||
| - | <div class="center" ><div class="thumb tnone" style="width: | + | <div class="center"><div class="thumb tnone" style="width:562px;"><div class="thumbinner" style="width:562px;"><a href="/File:Q().jpg" class="image"><img alt="Q().jpg" src="/wiki/images/thumb/e/e5/Q%28%29.jpg/560px-Q%28%29.jpg" width="560" height="220" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Q().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure4.Theoretical design 1</div></div></div></div> |
</html> | </html> | ||
| - | </ | + | <p align=center class="ppp"></p> |
| + | <p> In this design, the rhlPR is a RHLR/RHL Inducible Promoter, and we can assume the AI comes from pathogen is RHL. So the reprogrammed bacteria with green fluorescence will move toward the the infection site with a high concentration of RHL, while the reprogrammed bacteria with red fluorescence will stay at the original site waiting to creat the second wave of the reprogrammed bacteria with green fluorescence , then the reprogrammed bacteria with green fluorescence will decompose and release the pyosin to destroy the pathogens.</p> | ||
| + | <p>'''2.Other small molecules as chemical signals'''</p> | ||
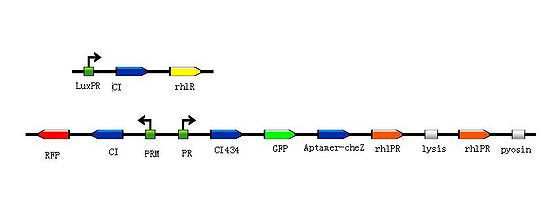
| + | <p> By using the device from the article ''Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen'' (Figure 3.), we can modify our original design to achieve the function which we have described before(Figure 5.).</p> | ||
| + | <html> | ||
| + | <div class="center"><div class="thumb tnone" style="width:562px;"><div class="thumbinner" style="width:562px;"><a href="/File:J().jpg" class="image"><img alt="J().jpg" src="/wiki/images/thumb/f/f8/J%28%29.jpg/560px-J%28%29.jpg" width="560" height="179" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:J().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure5.Theoretical design 2</div></div></div></div> | ||
| + | </html> | ||
| + | <p align=center class="ppp"></p> | ||
| + | <p> In this design, we assume the small molecule is theophylline. So the reprogrammed bacteria with green fluorescence will move toward the infection site with a high concentration of theophylline and the expression of Lysis becomes more and more, while the reprogrammed bacteria with red fluorescence will stay at the original site and wait to creat the second wave of the reprogrammed bacteria with green fluorescence, they have been protected from being destroyed by the pyosin from the reprogrammed bacteria with green fluorescence. The reprogrammed bacteria with green fluorescence will destroy themselves and release the Pyosin, which is expressed at the beginning of the process, to kill the pathogens.</p> | ||
| + | <p> Besides, the Lysis protein can be replaced by ccdB protein, and the Pyosin also can be substituted by other proteins of destruction, and the theophylline also can be replaced by other small molecules or catalyzed from other molecules like caffein or amino acid.</p> | ||
| + | <html><a name="Expermental Design and Results" id=""></a></html> | ||
| + | ==Simulated Experiment Design and Results <html><a href= "https://2011.igem.org/Team:USTC-China/Wet_Lab/protocol#Artificial"><font size="4"><u>(protocol)</u></font></a> </html>== | ||
| - | |||
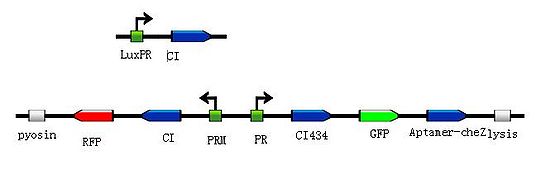
| - | + | <p> To put the original Toggle Switch-Aptamer-cheZ in use, we choose the second design above(As shown in Figure 5.) to be the simulated experiment construction(Figure 6.).</P> | |
<html> | <html> | ||
| - | <div class="center"><div class="thumb tnone" style="width: | + | <div class="center"><div class="thumb tnone" style="width:562px;"><div class="thumbinner" style="width:562px;"><a href="/File:G().jpg" class="image"><img alt="G().jpg" src="/wiki/images/thumb/e/ee/G%28%29.jpg/560px-G%28%29.jpg" width="560" height="171" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:G().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure6.Simulated experiment construction</div></div></div></div> |
</html> | </html> | ||
| - | + | <p align=center class="ppp"></p> | |
| - | < | + | <p> In this design, we use biobrick [http://partsregistry.org/Part:BBa_K117000 BBa_K117000] as the Lysis protein, and use biobrick [http://partsregistry.org/Part:BBa_K117001 BBa_K117001] to replace the pyosin. In which, the biobrick [http://partsregistry.org/Part:BBa_K117001 BBa_K117001] is ColE7-ImmE7 protein complex, and the lysis protein can remove Immunity protein from the complex with ColE7, then ColE7 regains its toxicity and ability to kill the EHEC bacteria or other bacteria nearby. Lysis protein itself can induce the lysis of host cell, exposing the interior cell contents and hence allows free diffusion of ColE7 proteins towards pathogenic bacteria strain.</P> |
| - | + | <p> Under the consideration of bio-safety, we just use the ordinary E.coli strain with the chloramphenicol and tetracycline resistance as the target bacteia. After the experiment (details are shown in the Protocol), we have some reliable results shown below.</P> | |
<html> | <html> | ||
| - | <div class="center"><div class="thumb tnone" style="width: | + | <div class="center"><div class="thumb tnone" style="width:314px;"><div class="thumbinner" style="width:314px;"><a href="/File:Nt().jpg" class="image"><img alt="Nt().jpg" src="/wiki/images/9/99/Nt%28%29.jpg" width="312" height="174" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Nt().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure7.The fluctuation of expression of the Toggle Switch device, the number of bacteria with red fluorescent decreases from tube1 to tube2.</div></div></div></div> |
</html> | </html> | ||
| + | <p align=center class="ppp"></p> | ||
| + | <html> | ||
| + | <div class="center"><div class="thumb tnone" style="width:652px;"><div class="thumbinner" style="width:652px;"><a href="/File:Zzy().jpg" class="image"><img alt="Zzy().jpg" src="/wiki/images/thumb/c/c5/Zzy%28%29.jpg/650px-Zzy%28%29.jpg" width="650" height="192" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Zzy().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure8.The growing state of the target bacteria. Left(after 3h), Right(after 10h)</div></div></div></div> | ||
| + | </html> | ||
| + | <p align=center class="ppp"></p> | ||
| + | <p> Figure8 shows the growing state of the target bacteria as the control of the experiment, in which the red circle represent the range of motion of the target, and from left to right the concentration of the theophylline increases.</P> | ||
| + | <html> | ||
| + | <div class="center"><div class="thumb tnone" style="width:652px;"><div class="thumbinner" style="width:652px;"><a href="/File:Qpp().jpg" class="image"><img alt="Qpp().jpg" src="/wiki/images/thumb/3/30/Qpp%28%29.jpg/650px-Qpp%28%29.jpg" width="650" height="184" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Qpp().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure9.The growing state of the reprogrammed bacteria(colony1,from tube2) and the target bacteria(colony2). Left(after 3h), Right(after 10h)</div></div></div></div> | ||
| + | </html> | ||
| + | <p align=center></p> | ||
| + | <P> Figure9 shows the growing state of the reprogrammed bacteria(from tube2) and the target bacteria, in which the yellow circle represent the range of motion of reprogrammed bacteria and the red circle represent the range of motion of the target bacteria, and from left to right the concentration of the theophylline increases.</p> | ||
| + | <html> | ||
| + | <div class="center"><div class="thumb tnone" style="width:652px;"><div class="thumbinner" style="width:652px;"><a href="/File:Aaa().jpg" class="image"><img alt="Aaa().jpg" src="/wiki/images/thumb/2/2b/Aaa%28%29.jpg/650px-Aaa%28%29.jpg" width="650" height="253" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Aaa().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure10.The fluorescent Photo(4X Objective)of two colonies shown in Figure9, the reprogrammed bacteria(Left,from tube2) and the target bacteria(Right).</div></div></div></div> | ||
| + | </html> | ||
| + | <p align=center class="ppp"></p> | ||
| + | <p> Figure10 shows two colonies, the colony of the reprogrammed bacteria which keeps the normal form, and the colony of the target bacteria which is deformed and has a hole as a plaque in the middle of it.</p> | ||
| + | | ||
| + | <html> | ||
| + | <div class="center"><div class="thumb tnone" style="width:352px;"><div class="thumbinner" style="width:352px;"><a href="/File:Ww().jpg" class="image"><img alt="Ww().jpg" src="/wiki/images/thumb/2/26/Ww%28%29.jpg/350px-Ww%28%29.jpg" width="350" height="298" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Ww().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure11.The growing state of the reprogrammed bacteria(colony1,from tube3) and the target bacteria(colony2)(after 10h)</div></div></div></div> | ||
| + | </html> | ||
| + | <p align=center class="ppp"></p> | ||
| + | <P> Figure11 shows the growing state of the reprogrammed bacteria(from tube3) and the target bacteria, in which the yellow circle represent the range of motion of reprogrammed bacteria and the red circle represent the range of motion of the target bacteria, and from left to right the concentration of the theophylline increases.</p> | ||
| + | <html> | ||
| + | <div class="center"><div class="thumb tnone" style="width:652px;"><div class="thumbinner" style="width:652px;"><a href="/File:Ddd().jpg" class="image"><img alt="Ddd().jpg" src="/wiki/images/thumb/a/a7/Ddd%28%29.jpg/650px-Ddd%28%29.jpg" width="650" height="236" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Ddd().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure12.The fluorescent Photo(4X Objective) of two colonies shown in Figure11, the reprogrammed bacteria(Left,from tube2) and the target bacteria(Right).</div></div></div></div> | ||
| + | </html> | ||
| + | <p align=center class="ppp"></p> | ||
| + | <p> Figure12 shows two colonies, the colony of the reprogrammed bacteria and the colony of the target bacteria,both of them are deformed and have holes as plaques in the middle of the colony.</p> | ||
| + | <html><a name="Conclusion and Discussion" id=""></a></html> | ||
| - | < | + | ==Conclusion and Discussion== |
| - | < | + | <p> Comparing with the control, the reprogrammed bacteria actually can destroy the target bacteria as the design requires and basically decrease the range of motion of the target bacteria.</p> |
| - | + | <p> By measuring the range of motion of the control and the target bacteria dealed with the reprogrammed bacteria from different tubes(tube1, tube2, tube3, tube4), we get histogram below.</p> | |
| - | + | ||
<html> | <html> | ||
| - | <div class="center"><div class="thumb tnone" style="width: | + | <div class="center"><div class="thumb tnone" style="width:502px;"><div class="thumbinner" style="width:502px;"><a href="/File:Wmm().jpg" class="image"><img alt="Wmm().jpg" src="/wiki/images/thumb/7/73/Wmm%28%29.jpg/500px-Wmm%28%29.jpg" width="500" height="255" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Wmm().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>;Figure13 shows that the capability of eliminating the target bacteria grows with the number of bacteria with red fluorescen decreases. This result reveal a fact that the properties of the reprogrammed bacteria mainly depend on the random fluctuation of the expression of the Toggle Switch Device.This fact also explain the differences between the form of the colony1 from tube2(Figure10 left) and the colony1 from tube3(Figure12 Left).</div></div></div></div> |
</html> | </html> | ||
| - | <p> | + | <p>  </p> |
| + | <p> Therefor to increase the controllability of this device, we can introduce the quorum sensing into our design to make the reprogrammed bacteria destroy the target more effectively.</P> | ||
| + | ==Future Work== | ||
| + | <p> The clinical potential of the application of the synthetic biology is very huge. Such potential includes the development of the therapies for the treatment of infectious diseases and cancer, as well as approaches in vaccine development, microbiome engineering, cell therapy, and regenerative medicine.</p> | ||
| + | <p> With such background, our Toggle Switch-Aptamer-cheZ Device show us a new way to fight against different diseases.Using our analysis result about [https://2011.igem.org/Team:USTC-China/Drylab/riboswitch Riboswitch], we can standardize the riboswitches or aptamers as Plug-ins ,then our device is just like a kind of "weapon" platform, which uses different standard equipments, such as bacteriocin(just like our design), RNAi(shown in Figure14), or some other metabolites, to cure the diseases and keep us healthy.</p> | ||
<html> | <html> | ||
| - | <div class="center"><div class="thumb tnone" style="width: | + | <div class="center"><div class="thumb tnone" style="width:470px;"><div class="thumbinner" style="width:470px;"><a href="/File:Wdm().jpg" class="image"><img alt="Wdm().jpg" src="/wiki/images/2/28/Wdm%28%29.jpg" width="468" height="199" class="thumbimage" /></a> <div class="thumbcaption"><div class="magnify"><a href="/File:Wdm().jpg" class="internal" title="Enlarge"><img src="/wiki/skins/common/images/magnify-clip.png" width="15" height="11" alt="" /></a></div>Figure14.This image comes from Warren C. Ruder.etc(2011)</div></div></div></div> |
</html> | </html> | ||
| - | <p> | + | <p align=center class="ppp"></p> |
| + | |||
| + | ==References== | ||
| + | <p>1.Timothy S. Gardner, Charles R. Cantor, James J. Collins (2000) Construction of a | ||
| + | Genetic toggle switch in Escherichia coli. Nature, 403,339-342</p> | ||
| + | <p>2.Shana Topp, Justin P. Gallivan (2006) Guiding Bacteria with Small Molecules and RNA. J.Am.Chem.Soc, 129,6870-6811</p> | ||
| + | <p>3.Chunbo Lou, Xili Liu, Ming Ni.etc (2010) Synthesizing a novel genetic sequential logic circuit: a push-on push-off switch. Molecular Systems Biology, 6:350</p> | ||
| + | <p>4.Joy Sinha, Samuel J Reyes, Justin P Gallivan (2010) Reprogramming bacteria to seek and destroy an herbicide. Nature Chemical Biology, 6,464-470</p> | ||
| + | <p>5.Steven L. Porter, George H. Wadhams, Judith P. Armitage (2011) Signal processing in complex chemotaxis pathways. Nature Reviews Microbiology, 9,153-165</p> | ||
| + | <p>6.Warren C. Ruder, Ting Lu, James J. Collins (2011) Synthetic Biology Moving into the Clinic. Science,333,1248-1252</p> | ||
| + | <p>7.Nazanin Saeidi, Choon Kit Wong, Tat-Ming Lo .etc (2011) Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Molecular Systems Biology,7:251</p> | ||
Latest revision as of 13:31, 27 October 2011

Contents |
Artificial Innate Immunity System(AIIS)
1.Background
2.Purpose
3.Principle
4.Theoretical Design
5.Expermental Design and Results
6.Conclusion and Discussion
Background
From mid May to June 2011, enteropathogenic E.coli brings a big panic to many different European countries. Why such an ordinary bacteria as E.coli can lead to a clinical catastrophe and kill a lot of people? The answer may be complicate, but the most important reason is that we can not find out the pathogens rapidly and can not apply the effective treatments.To solve this problem, we try to use the normal E.coli, which keep the symbiotic relationship with us humanbeings, to be a safety weapon to defend ourselves against pathogens.
Purpose
Using reprogrammed intestinal bacteria as a safety weapon to destroy the pathogens which invade the intestinal mucosal system.
Principle
As shown in Figure2, When pathogens invade the intestinal epithelium ,they will release some kind of chemical signal. After the reprogrammed intestinal bacteria capture this signal, such signal can activate the expression of the function gene ,then it will drive the reprogrammed intestinal bacteria to move toward the infection site from the original colony. Then the moving reprogrammed bacteria will express some lethal proteins at the infection site to kill pathogens by self-destruction, and the original colony will remain.
Theoretical Design
1.AI(Auto Inducer)as a Chemical Signal
By using the device from the article Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen (Figure3.), we can modify our original design to achieve the function which we have described before.(As shown in Figure4.)
In this design, the rhlPR is a RHLR/RHL Inducible Promoter, and we can assume the AI comes from pathogen is RHL. So the reprogrammed bacteria with green fluorescence will move toward the the infection site with a high concentration of RHL, while the reprogrammed bacteria with red fluorescence will stay at the original site waiting to creat the second wave of the reprogrammed bacteria with green fluorescence , then the reprogrammed bacteria with green fluorescence will decompose and release the pyosin to destroy the pathogens.
2.Other small molecules as chemical signals
By using the device from the article Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen (Figure 3.), we can modify our original design to achieve the function which we have described before(Figure 5.).
In this design, we assume the small molecule is theophylline. So the reprogrammed bacteria with green fluorescence will move toward the infection site with a high concentration of theophylline and the expression of Lysis becomes more and more, while the reprogrammed bacteria with red fluorescence will stay at the original site and wait to creat the second wave of the reprogrammed bacteria with green fluorescence, they have been protected from being destroyed by the pyosin from the reprogrammed bacteria with green fluorescence. The reprogrammed bacteria with green fluorescence will destroy themselves and release the Pyosin, which is expressed at the beginning of the process, to kill the pathogens.
Besides, the Lysis protein can be replaced by ccdB protein, and the Pyosin also can be substituted by other proteins of destruction, and the theophylline also can be replaced by other small molecules or catalyzed from other molecules like caffein or amino acid.
Simulated Experiment Design and Results (protocol)
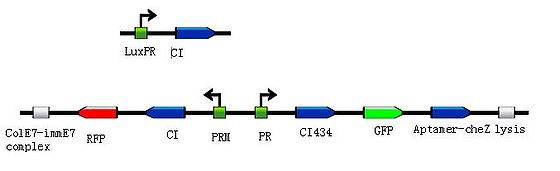
To put the original Toggle Switch-Aptamer-cheZ in use, we choose the second design above(As shown in Figure 5.) to be the simulated experiment construction(Figure 6.).
In this design, we use biobrick [http://partsregistry.org/Part:BBa_K117000 BBa_K117000] as the Lysis protein, and use biobrick [http://partsregistry.org/Part:BBa_K117001 BBa_K117001] to replace the pyosin. In which, the biobrick [http://partsregistry.org/Part:BBa_K117001 BBa_K117001] is ColE7-ImmE7 protein complex, and the lysis protein can remove Immunity protein from the complex with ColE7, then ColE7 regains its toxicity and ability to kill the EHEC bacteria or other bacteria nearby. Lysis protein itself can induce the lysis of host cell, exposing the interior cell contents and hence allows free diffusion of ColE7 proteins towards pathogenic bacteria strain.
Under the consideration of bio-safety, we just use the ordinary E.coli strain with the chloramphenicol and tetracycline resistance as the target bacteia. After the experiment (details are shown in the Protocol), we have some reliable results shown below.
Figure8 shows the growing state of the target bacteria as the control of the experiment, in which the red circle represent the range of motion of the target, and from left to right the concentration of the theophylline increases.
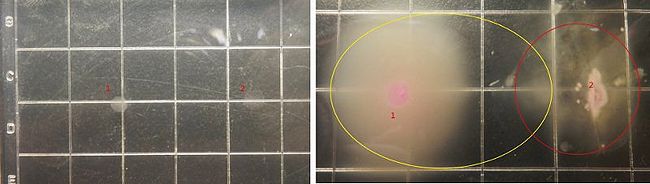
Figure9 shows the growing state of the reprogrammed bacteria(from tube2) and the target bacteria, in which the yellow circle represent the range of motion of reprogrammed bacteria and the red circle represent the range of motion of the target bacteria, and from left to right the concentration of the theophylline increases.
Figure10 shows two colonies, the colony of the reprogrammed bacteria which keeps the normal form, and the colony of the target bacteria which is deformed and has a hole as a plaque in the middle of it.
Figure11 shows the growing state of the reprogrammed bacteria(from tube3) and the target bacteria, in which the yellow circle represent the range of motion of reprogrammed bacteria and the red circle represent the range of motion of the target bacteria, and from left to right the concentration of the theophylline increases.
Figure12 shows two colonies, the colony of the reprogrammed bacteria and the colony of the target bacteria,both of them are deformed and have holes as plaques in the middle of the colony.
Conclusion and Discussion
Comparing with the control, the reprogrammed bacteria actually can destroy the target bacteria as the design requires and basically decrease the range of motion of the target bacteria.
By measuring the range of motion of the control and the target bacteria dealed with the reprogrammed bacteria from different tubes(tube1, tube2, tube3, tube4), we get histogram below.
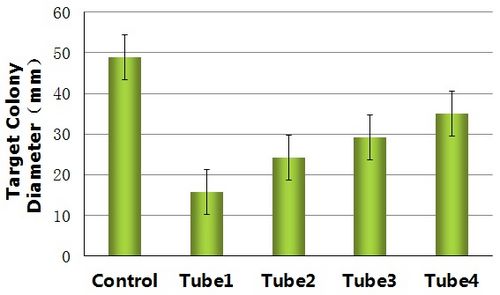
 
Therefor to increase the controllability of this device, we can introduce the quorum sensing into our design to make the reprogrammed bacteria destroy the target more effectively.
Future Work
The clinical potential of the application of the synthetic biology is very huge. Such potential includes the development of the therapies for the treatment of infectious diseases and cancer, as well as approaches in vaccine development, microbiome engineering, cell therapy, and regenerative medicine.
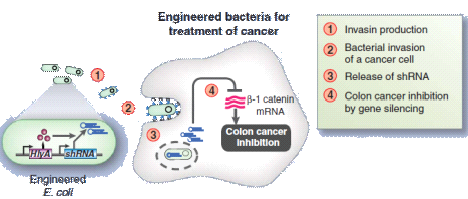
With such background, our Toggle Switch-Aptamer-cheZ Device show us a new way to fight against different diseases.Using our analysis result about Riboswitch, we can standardize the riboswitches or aptamers as Plug-ins ,then our device is just like a kind of "weapon" platform, which uses different standard equipments, such as bacteriocin(just like our design), RNAi(shown in Figure14), or some other metabolites, to cure the diseases and keep us healthy.
References
1.Timothy S. Gardner, Charles R. Cantor, James J. Collins (2000) Construction of a Genetic toggle switch in Escherichia coli. Nature, 403,339-342
2.Shana Topp, Justin P. Gallivan (2006) Guiding Bacteria with Small Molecules and RNA. J.Am.Chem.Soc, 129,6870-6811
3.Chunbo Lou, Xili Liu, Ming Ni.etc (2010) Synthesizing a novel genetic sequential logic circuit: a push-on push-off switch. Molecular Systems Biology, 6:350
4.Joy Sinha, Samuel J Reyes, Justin P Gallivan (2010) Reprogramming bacteria to seek and destroy an herbicide. Nature Chemical Biology, 6,464-470
5.Steven L. Porter, George H. Wadhams, Judith P. Armitage (2011) Signal processing in complex chemotaxis pathways. Nature Reviews Microbiology, 9,153-165
6.Warren C. Ruder, Ting Lu, James J. Collins (2011) Synthetic Biology Moving into the Clinic. Science,333,1248-1252
7.Nazanin Saeidi, Choon Kit Wong, Tat-Ming Lo .etc (2011) Engineering microbes to sense and eradicate Pseudomonas aeruginosa, a human pathogen. Molecular Systems Biology,7:251
 "
"