Team:TzuChiU Formosa/Project/PhotoPaper
From 2011.igem.org
| (41 intermediate revisions not shown) | |||
| Line 37: | Line 37: | ||
<br><font size=4><b>a. about cellulose biosynthesis</b></font> | <br><font size=4><b>a. about cellulose biosynthesis</b></font> | ||
<br>Bacterial cellulose is a form of cellulose which is produced by bacteria. It has the same molecular formula as green plants, which is a polysaccharide with thousands of β(1→4) linked D-glucose units. Cellulose biosynthesis is a carbohydrate metabolism with several enzyme regulations, and the key factor of this pathway is cellulose synthase, which participates in the transition from UDP-glucose to cellulose. The ligand of cellulose synthase is cyclic di-GMP, an effector forming from 2 molecules of GTP. The enzyme is activated by the binding of c-di-GMP with cellulose synthase, turning UDP-glucose into UDP, meanwhile form β-1,4-glucan chains, which are then used to synthesize cellulose. | <br>Bacterial cellulose is a form of cellulose which is produced by bacteria. It has the same molecular formula as green plants, which is a polysaccharide with thousands of β(1→4) linked D-glucose units. Cellulose biosynthesis is a carbohydrate metabolism with several enzyme regulations, and the key factor of this pathway is cellulose synthase, which participates in the transition from UDP-glucose to cellulose. The ligand of cellulose synthase is cyclic di-GMP, an effector forming from 2 molecules of GTP. The enzyme is activated by the binding of c-di-GMP with cellulose synthase, turning UDP-glucose into UDP, meanwhile form β-1,4-glucan chains, which are then used to synthesize cellulose. | ||
| + | <br> | ||
| + | <html> | ||
| + | <object type="application/x-shockwave-flash" height="800" width="600" data="https://static.igem.org/mediawiki/2011/6/61/Cellulose_biosynthesis.swf"> | ||
| + | <param name="movie" value="https://static.igem.org/mediawiki/2011/6/61/Cellulose_biosynthesis.swf" /> | ||
| + | <param name="quality" value="high" /> | ||
| + | <param name="wmode" value="transparent"> | ||
| + | |||
| + | </object> | ||
| + | </html> | ||
| + | |||
<br> | <br> | ||
<br><font size=4><b>b.about acs operon</b></font> | <br><font size=4><b>b.about acs operon</b></font> | ||
<br> | <br> | ||
| + | [[File:Screen shot 2011-10-06 at 上午5.45.47.png|right|350px]] | ||
<i>Gluconacetobacter hansenii</i> ATCC23769 has been characterized as a model organism for cellulose biosynthesis. The cluster of genes which play the role of producing cellulose is acs operon. It contains three major genes, acsAB, acsC and acsD, and the final product is cellulose synthase. The acsAB catalyzes the formation of bacterial cellulose; acsA is the catalytic subunit which utilizes UDP-glucose to form the basic unit of cellulose; while acsB provide the regulatory subunit which has a cyclic di-GMP binding domain. The acsC gene is the main composite in the formation of the membrane complex of cellulose synthase and proposed to be involved in the export of the polymer across the bacterial cell wall.The acsD gene is involved in the crystallization of the mature cellulose by cleaves the intrastrand ß-1,4 linkages in the cellulose chain and is proposed to have a role in the release of the growing polymer from the cell. | <i>Gluconacetobacter hansenii</i> ATCC23769 has been characterized as a model organism for cellulose biosynthesis. The cluster of genes which play the role of producing cellulose is acs operon. It contains three major genes, acsAB, acsC and acsD, and the final product is cellulose synthase. The acsAB catalyzes the formation of bacterial cellulose; acsA is the catalytic subunit which utilizes UDP-glucose to form the basic unit of cellulose; while acsB provide the regulatory subunit which has a cyclic di-GMP binding domain. The acsC gene is the main composite in the formation of the membrane complex of cellulose synthase and proposed to be involved in the export of the polymer across the bacterial cell wall.The acsD gene is involved in the crystallization of the mature cellulose by cleaves the intrastrand ß-1,4 linkages in the cellulose chain and is proposed to have a role in the release of the growing polymer from the cell. | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br><br><font color="#000080" size=5>System Design</font><br> | ||
| - | |||
<Hr Align="left" width="100%" size=2> | <Hr Align="left" width="100%" size=2> | ||
<br> | <br> | ||
| + | <font size = 4><b>1.Biobricks</b></font><br><br> | ||
| + | Because of the long sequence of complete acs operon, we designed several pairs of | ||
| + | primers to amplify acsAB and acsCD, the products are then purified and digested with | ||
| + | EcoRI and SpeI, next ligated with the pSB1C3 cut by EcoRI-SpeI, which pSB1C3 is the | ||
| + | iGEM 2011 standard backbone. | ||
| + | [[File:Screen shot 2011-10-06 at 上午10.32.16.png|center|600px]] | ||
| + | <br> | ||
| + | <br> | ||
| + | <font size = 4><b>2. Expression</b></font><br><br> | ||
| + | |||
| + | The recombinant plasmids would be introduced into <i>E. coli</i> for testing and modelling | ||
| + | before transforming into <i>R. rubrum.</i> The IPTG-induced promoter is cloned into biobricks so | ||
| + | that the acs operon expression would be regulated by the addition of IPTG. | ||
| + | As to <i>R. rubrum</i>, the recombinant plasmids would be introduced by electroporation. The | ||
| + | glucose which produced by photosynthesis would be the raw material for cellulose | ||
| + | synthesis. | ||
| + | <br> | ||
| + | <br> | ||
| + | <font size = 4><b>3. Modelling | ||
| + | </b></font><br><br> | ||
| + | Observation of the performance of acs ABCD protein during PAGE can predict the | ||
| + | productivity of cellulose, run 2 SDS-PAGE, one according to the reaction time and another | ||
| + | according to the presence of promoter. | ||
| + | The bacterial culture which have been induced were collected and the bacteria is to | ||
| + | remove, the end product is added with cellulase until the cellulose is fully decomposed. | ||
| + | Benedict solution caused the formation of brick red precipitate, the concentration of | ||
| + | cellulose is then tested with OD645. | ||
| + | [[File:Cellulose.jpg|center|700px]] | ||
| + | <html> | ||
| + | <object type="application/x-shockwave-flash" height="400" width="800" data="https://static.igem.org/mediawiki/2011/3/39/Overview.swf"> | ||
| + | <param name="movie" value="https://static.igem.org/mediawiki/2011/3/39/Overview.swf" /> | ||
| + | <param name="quality" value="high" /> | ||
| + | <param name="wmode" value="transparent"> | ||
| + | </object> | ||
| + | </html> | ||
<br><br><font color="#000080" size=5>Result</font> | <br><br><font color="#000080" size=5>Result</font> | ||
<Hr Align="left" width="100%" size=2> | <Hr Align="left" width="100%" size=2> | ||
| + | |||
<br> | <br> | ||
| - | + | Every wanted gene is successfully cloned. Beside the anticipated parts, we also | |
| - | + | constructed other biobricks which carry more insert genes. Transformation of every | |
| + | biobricks in E. coli is accomplished too. | ||
| + | |||
| + | [[File:Screen shot 2011-10-06 at 上午11.12.32.png|400px]] | ||
| + | [[File:Screen shot 2011-10-06 at 上午11.16.31.png|400px]] | ||
<br> | <br> | ||
| - | = | + | <br> |
| - | [[ | + | <br> |
| + | Gluconacetobacter hansenii which carries the acsABCD protein has the ability to produce cellulose is our main character in our project. Our project is to transform acsABCD genes into E. coli to monitor the production of cellulose production. We collect the protein produced by the transformed E.coli and Gluconacetobacter hansenii in different time point to monitor the expression of acsABCD gene products and the production of cellulose. | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <font size=3><b>Method 1 : SDS-PAGE</b></font> | ||
| + | <br> | ||
| + | <br> | ||
| + | Observe the performance of acs ABCD protein during PAGE to predict the productivity of cellulose, run 2 SDS-PAGE respectively, one according to the reaction time and another according to the presence of promoter. | ||
| + | <br> | ||
| + | <br> | ||
| + | The protein purification from <i>E.Coli</i> with acsAB and with acsCD is use to run the SDS-PAGE in order to compare the expression of every protein sample. The expression is then use to predict and calculate the amount of protein produced by the <i>Gluconacetobacter hansenii</i>. | ||
| + | <br> | ||
| + | <br> | ||
| + | The SDS-PAGE is run twice. One is with promoter and another one without promoter. The difference of the protein produced is used to predict productivity of cellulose. At 0 hr, the <i>E. coli</i> which carries promoter ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K571004 BBa_k571004]) + acsAB or promoter ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K571004 BBa_k571004]) + acsCD all performed well. At 4hrs, the gene expression is shown as well, especially the gene which carries promoter+acsCD. The gene which carries promoter+acsCD expressed constantly until 10hrs. | ||
| + | <br> | ||
| + | <br> | ||
| + | The relative molecular mass of acsAB is about 168KDa, while the relative molecular mass of acsCD is about 155KDa. Therefore, we predict that the protein expression should be somewhere between this 2 bands. Therefore, we can conclude that the primer we designed is functional. | ||
| + | <br> | ||
| + | 1. Compare the positive control with acsAB gene and acsCD gene | ||
| + | <br> | ||
| + | 2. Calculate the amount of protein produced | ||
| + | <br> | ||
| + | 3. Predict the productivity of cellulose | ||
| + | <br> | ||
| + | '''-difference of promoter''' | ||
| + | <br>[[File:Pro.jpg|center|800px]] | ||
| + | <br> | ||
| + | <br> | ||
| - | + | <br> | |
| - | < | + | '''-difference of reaction time''' |
| - | <object type="application/x-shockwave-flash" height=" | + | <br> |
| - | <param name="movie" value="https://static.igem.org/mediawiki/2011/3/ | + | 0hr, 4hrs, 8hrs, 10hrs, and 12hrs of bacterial culture were collected. Compare the high-performance phase of the acs ABCD protein, calculate the productivity of cellulose. |
| + | |||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <font size=3><b>Method 2 : Benedict’s test</b></font> | ||
| + | <br> | ||
| + | <br> | ||
| + | The bacterial culture which have been induced were collected and the bacteria is to remove, the end product is added with cellulase until the cellulose is fully decomposed. Benedict solution caused the formation of brick red precipitate, the concentration of cellulose is then tested with OD645. | ||
| + | <br> | ||
| + | <br> | ||
| + | Cellulase is used to break down the cellulose into monosaccharide. Cellulose is a glucose polymer connected through a beta (1-4) glycosidic linkages. Benedict’s test is carried out to test the presence of reducing sugar, such as. The reducing sugar reduces copper(II) ions in these test solutions to copper(I), which then forms a brick red copper(I) oxide precipitate. The color would range from green to brick red respectively depends on the amount of reducing sugar present in the solution. It can detect the concentration of the reducing sugar under the absorbance condition <sub>OD645</sub>. | ||
| + | |||
| + | Cellulose (beta-1,4 glucan) is the most plentiful biopolymer in nature and is an crucial raw material for many industries. It is synthesized as extracellular fibrils by cellulose synthase not only in plants but also in some bacteria. | ||
| + | |||
| + | Bacteria with cellulose synthase gene use Isopropyl-β- D -1-thiogalactopyranoside (IPTG) as the inducer stimulating the production of protein. In our system, IPTG act as the inducer of the R0011 promoter, which then activate the operon. The activated acs operon encodes the cellulose synthase to synthesize cellulose. The reaction lasts two hours and every two hour we would need to collect the purified cellulose. The nett weight is recorded and the mechanism of cellulose activity on pure cellulose substrates is identified. Lastly, the Benedict’s solution is added to find out the absorbance (optical density [O.D.]) value, in order to calculate the amount of monosaccharide that cellulose can produce. | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <html><object type="application/x-shockwave-flash" height="450" width=650" data="https://static.igem.org/mediawiki/2011/3/3e/Growth_curve.swf"> | ||
| + | <param name="movie" value="https://static.igem.org/mediawiki/2011/3/3e/Growth_curve.swf" /> | ||
<param name="quality" value="high" /> | <param name="quality" value="high" /> | ||
<param name="wmode" value="transparent"> | <param name="wmode" value="transparent"> | ||
</object> | </object> | ||
</html> | </html> | ||
| + | |||
| + | '''Bacterial cellulose''' | ||
| + | <div align ="center"> | ||
| + | [[File:Big1.JPG|300px]][[File:Big2.JPG|300px]]</div> | ||
| + | |||
| + | <br><html><div align="center">the sample of bacterial cellulose that we collected in 3 days</div></html> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | <br> | ||
| + | [[File:111456.jpg|center|500px]] | ||
| + | <br><html><div align="center">bacterial cellulose under EM<br><br><br> | ||
| + | </div> | ||
| + | <br> | ||
| + | <br><br><font color="#000080" size=5>References</font> | ||
| + | <Hr Align="left" width="100%" size=2> | ||
| + | <br> | ||
| + | 1.Rune Standal, Tore-geir Iversen, Dag H. Coucheron, Espen Fjaervik, Janet M. Blantny, Svein Valla. A New Gene Required for Cellulose Production and a Gen Encoding Cellulolytic Activity in Acetobacter xylinum Are Colocalized with the bcs operon. Journal of Bacteriology. Feb, 1994. | ||
| + | <br> | ||
| + | 2.Inder M. Saxena, Krystyna Kudlicka, Kazuo Okuda, R. Malcolm Brown, JR. Characterization of Genes in the Cellulose-Synthesizing Operon(acs operon) of Acetobacter xylinum: Implications for Cellulose Crystallization. Journal of Bacteriology. Sep,1994. | ||
| + | <br> | ||
| + | 3.Shin Kawano, Kenji Tajima, Yukako Uemori, Hitomi Yamashita, Tomoki Erata, Masanobu Munekata, Mitsuo Takai. DNA Research. October, 2002. | ||
| + | <br> | ||
| + | 4.Joel Weadge, Wilfrid Laurier University. | ||
Latest revision as of 03:57, 6 October 2011


Photopaper
Abstract
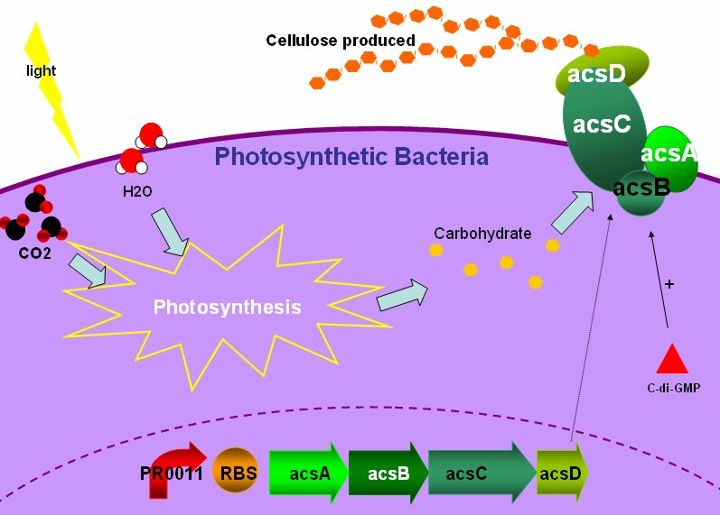
For most countries, paper-making has been a traditional but indispensable industry. Wood pulp is the major raw material for paper-making ,moreover,the complicated processes toward paper-making may contribute to environmental pollution. Acetobacter xylinum is a bacterium which produces bacterial cellulose. It has an acs operon, consisting of genes that called acsAB, acsC, and acsD. These genes interact with each other and synthesize cellulose synthase, an enzyme that transforms UDP-glucose into cellulose. What we want to do is to choose cyanobacteria which provides glucose through photosynthesis. Therefore, we want to use cyanobacteria as the host, then introduce the acs operon genes in it and produce bacterial cellulose by expressing this series of genes. With manufacturing processes, we believe this project can develop into a new and eco-friendly technology of papermaking.
Background
a. about cellulose biosynthesis
Bacterial cellulose is a form of cellulose which is produced by bacteria. It has the same molecular formula as green plants, which is a polysaccharide with thousands of β(1→4) linked D-glucose units. Cellulose biosynthesis is a carbohydrate metabolism with several enzyme regulations, and the key factor of this pathway is cellulose synthase, which participates in the transition from UDP-glucose to cellulose. The ligand of cellulose synthase is cyclic di-GMP, an effector forming from 2 molecules of GTP. The enzyme is activated by the binding of c-di-GMP with cellulose synthase, turning UDP-glucose into UDP, meanwhile form β-1,4-glucan chains, which are then used to synthesize cellulose.
b.about acs operon
Gluconacetobacter hansenii ATCC23769 has been characterized as a model organism for cellulose biosynthesis. The cluster of genes which play the role of producing cellulose is acs operon. It contains three major genes, acsAB, acsC and acsD, and the final product is cellulose synthase. The acsAB catalyzes the formation of bacterial cellulose; acsA is the catalytic subunit which utilizes UDP-glucose to form the basic unit of cellulose; while acsB provide the regulatory subunit which has a cyclic di-GMP binding domain. The acsC gene is the main composite in the formation of the membrane complex of cellulose synthase and proposed to be involved in the export of the polymer across the bacterial cell wall.The acsD gene is involved in the crystallization of the mature cellulose by cleaves the intrastrand ß-1,4 linkages in the cellulose chain and is proposed to have a role in the release of the growing polymer from the cell.
System Design
1.Biobricks
Because of the long sequence of complete acs operon, we designed several pairs of primers to amplify acsAB and acsCD, the products are then purified and digested with EcoRI and SpeI, next ligated with the pSB1C3 cut by EcoRI-SpeI, which pSB1C3 is the iGEM 2011 standard backbone.
2. Expression
The recombinant plasmids would be introduced into E. coli for testing and modelling
before transforming into R. rubrum. The IPTG-induced promoter is cloned into biobricks so
that the acs operon expression would be regulated by the addition of IPTG.
As to R. rubrum, the recombinant plasmids would be introduced by electroporation. The
glucose which produced by photosynthesis would be the raw material for cellulose
synthesis.
3. Modelling
Observation of the performance of acs ABCD protein during PAGE can predict the
productivity of cellulose, run 2 SDS-PAGE, one according to the reaction time and another
according to the presence of promoter.
The bacterial culture which have been induced were collected and the bacteria is to
remove, the end product is added with cellulase until the cellulose is fully decomposed.
Benedict solution caused the formation of brick red precipitate, the concentration of
cellulose is then tested with OD645.
Result
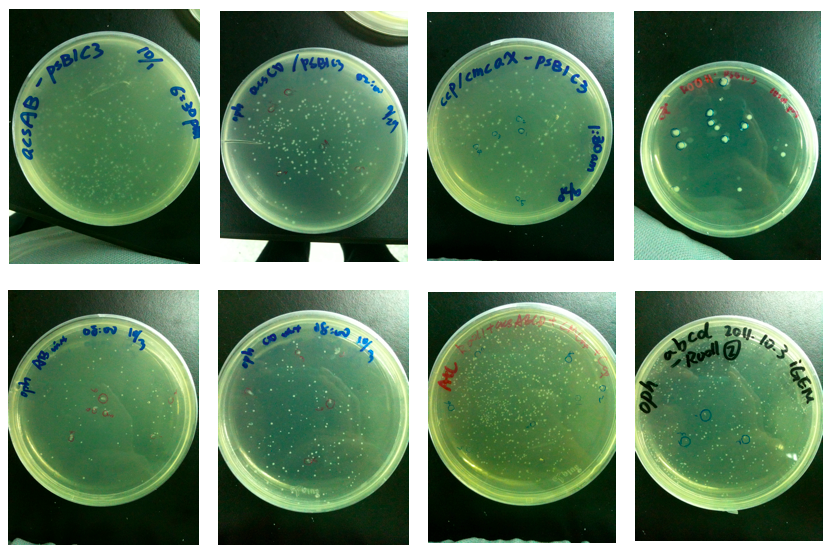
Every wanted gene is successfully cloned. Beside the anticipated parts, we also
constructed other biobricks which carry more insert genes. Transformation of every
biobricks in E. coli is accomplished too.

Gluconacetobacter hansenii which carries the acsABCD protein has the ability to produce cellulose is our main character in our project. Our project is to transform acsABCD genes into E. coli to monitor the production of cellulose production. We collect the protein produced by the transformed E.coli and Gluconacetobacter hansenii in different time point to monitor the expression of acsABCD gene products and the production of cellulose.
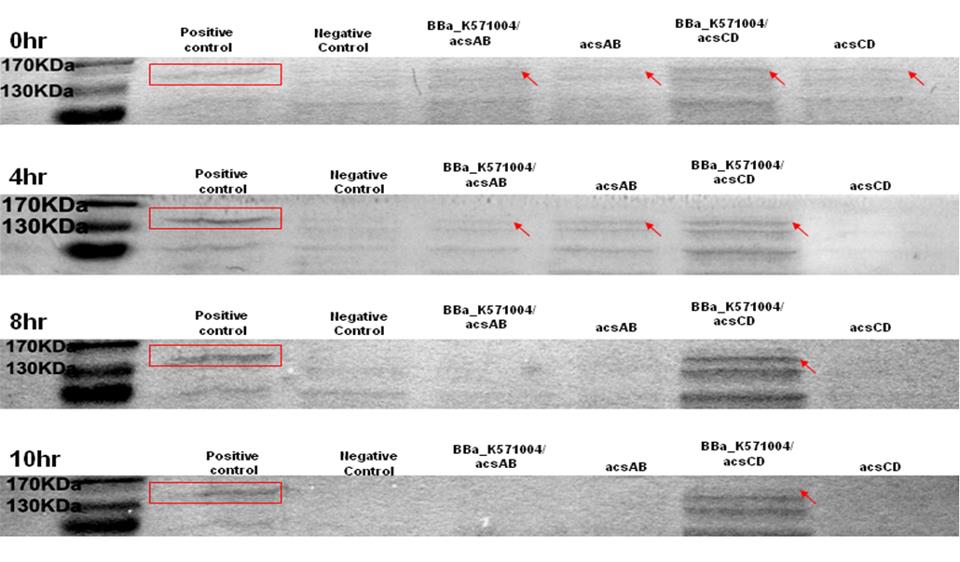
Method 1 : SDS-PAGE
Observe the performance of acs ABCD protein during PAGE to predict the productivity of cellulose, run 2 SDS-PAGE respectively, one according to the reaction time and another according to the presence of promoter.
The protein purification from E.Coli with acsAB and with acsCD is use to run the SDS-PAGE in order to compare the expression of every protein sample. The expression is then use to predict and calculate the amount of protein produced by the Gluconacetobacter hansenii.
The SDS-PAGE is run twice. One is with promoter and another one without promoter. The difference of the protein produced is used to predict productivity of cellulose. At 0 hr, the E. coli which carries promoter ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K571004 BBa_k571004]) + acsAB or promoter ([http://partsregistry.org/wiki/index.php?title=Part:BBa_K571004 BBa_k571004]) + acsCD all performed well. At 4hrs, the gene expression is shown as well, especially the gene which carries promoter+acsCD. The gene which carries promoter+acsCD expressed constantly until 10hrs.
The relative molecular mass of acsAB is about 168KDa, while the relative molecular mass of acsCD is about 155KDa. Therefore, we predict that the protein expression should be somewhere between this 2 bands. Therefore, we can conclude that the primer we designed is functional.
1. Compare the positive control with acsAB gene and acsCD gene
2. Calculate the amount of protein produced
3. Predict the productivity of cellulose
-difference of promoter
-difference of reaction time
0hr, 4hrs, 8hrs, 10hrs, and 12hrs of bacterial culture were collected. Compare the high-performance phase of the acs ABCD protein, calculate the productivity of cellulose.
Method 2 : Benedict’s test
The bacterial culture which have been induced were collected and the bacteria is to remove, the end product is added with cellulase until the cellulose is fully decomposed. Benedict solution caused the formation of brick red precipitate, the concentration of cellulose is then tested with OD645.
Cellulase is used to break down the cellulose into monosaccharide. Cellulose is a glucose polymer connected through a beta (1-4) glycosidic linkages. Benedict’s test is carried out to test the presence of reducing sugar, such as. The reducing sugar reduces copper(II) ions in these test solutions to copper(I), which then forms a brick red copper(I) oxide precipitate. The color would range from green to brick red respectively depends on the amount of reducing sugar present in the solution. It can detect the concentration of the reducing sugar under the absorbance condition OD645.
Cellulose (beta-1,4 glucan) is the most plentiful biopolymer in nature and is an crucial raw material for many industries. It is synthesized as extracellular fibrils by cellulose synthase not only in plants but also in some bacteria.
Bacteria with cellulose synthase gene use Isopropyl-β- D -1-thiogalactopyranoside (IPTG) as the inducer stimulating the production of protein. In our system, IPTG act as the inducer of the R0011 promoter, which then activate the operon. The activated acs operon encodes the cellulose synthase to synthesize cellulose. The reaction lasts two hours and every two hour we would need to collect the purified cellulose. The nett weight is recorded and the mechanism of cellulose activity on pure cellulose substrates is identified. Lastly, the Benedict’s solution is added to find out the absorbance (optical density [O.D.]) value, in order to calculate the amount of monosaccharide that cellulose can produce.
Bacterial cellulose
References
1.Rune Standal, Tore-geir Iversen, Dag H. Coucheron, Espen Fjaervik, Janet M. Blantny, Svein Valla. A New Gene Required for Cellulose Production and a Gen Encoding Cellulolytic Activity in Acetobacter xylinum Are Colocalized with the bcs operon. Journal of Bacteriology. Feb, 1994.
2.Inder M. Saxena, Krystyna Kudlicka, Kazuo Okuda, R. Malcolm Brown, JR. Characterization of Genes in the Cellulose-Synthesizing Operon(acs operon) of Acetobacter xylinum: Implications for Cellulose Crystallization. Journal of Bacteriology. Sep,1994.
3.Shin Kawano, Kenji Tajima, Yukako Uemori, Hitomi Yamashita, Tomoki Erata, Masanobu Munekata, Mitsuo Takai. DNA Research. October, 2002.
4.Joel Weadge, Wilfrid Laurier University.
 "
"