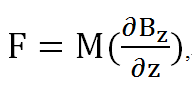Team:NYMU-Taipei/optomagnetic-design
From 2011.igem.org
(→Our Constructs) |
CHIACHENHSU (Talk | contribs) |
||
| Line 5: | Line 5: | ||
| - | ==Optomagnetic Design== | + | ==Optomagnetic Design==(by 尚叡) |
| - | (by 尚叡) | + | ==Magnetic Bacteria==(by 尚叡) |
| - | ==Magnetic Bacteria== | + | ==The Anchor Protein==(by 尚叡) |
| - | (by 尚叡) | + | |
| - | ==The Anchor Protein== | + | |
| - | (by 尚叡) | + | |
| - | + | ==The BRET Phenomenon== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | |
How to use the conformation change of the Mms13 transmembrane protein to induce the light? We first came up with the idea by using bioluminescence resonance energy transfer (BRET). BRET is based on resonance energy transfer between a light-emitting enzyme and a fluorescent acceptor. (Johan Bacart, et al., 2008) For example, the energy derived from a luciferase reaction can be used to excite a fluorescent protein when the latter is in close proximity to the luciferase enzyme. Several other methods developed are shown in Table 1. | How to use the conformation change of the Mms13 transmembrane protein to induce the light? We first came up with the idea by using bioluminescence resonance energy transfer (BRET). BRET is based on resonance energy transfer between a light-emitting enzyme and a fluorescent acceptor. (Johan Bacart, et al., 2008) For example, the energy derived from a luciferase reaction can be used to excite a fluorescent protein when the latter is in close proximity to the luciferase enzyme. Several other methods developed are shown in Table 1. | ||
| - | [[Image: table1_NYMU.png| | + | [[Image: table1_NYMU.png|frame|Table 1: Several BRET methods have been developed.( Cite from: http://www.berthold.com/ww/en/pub/bioanalytik/applikation/bret.cfm)]] |
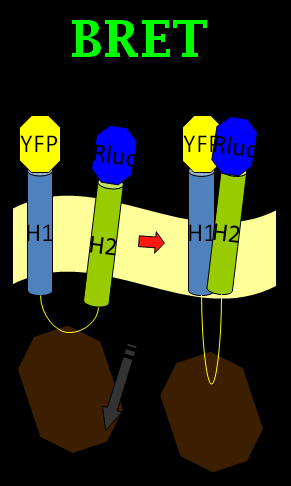
We chose the original BRET method using coelenterazine (luciferin) as substrate in BRET 1 method (See Figure 1). What we do is anchoring the YFP on the N-terminus of Mms13 and r-Luciferase on the C-terminus. We expect that when the magnetic force applied on the membrane and changed the conformation of Mms13 would lead to the close proximity between YFP and r-Luciferase and thus induce fluorescent phenomenon by BRET method (See Figure 1). | We chose the original BRET method using coelenterazine (luciferin) as substrate in BRET 1 method (See Figure 1). What we do is anchoring the YFP on the N-terminus of Mms13 and r-Luciferase on the C-terminus. We expect that when the magnetic force applied on the membrane and changed the conformation of Mms13 would lead to the close proximity between YFP and r-Luciferase and thus induce fluorescent phenomenon by BRET method (See Figure 1). | ||
| Line 29: | Line 21: | ||
The advantage of BRET is that it is a non-radioactive and homogeneous technology. There is no auto-fluorescence coming from compounds or cell and buffer components as no light source is required. And the most of all, it make the conformation change of Mms13 possible to induce optomagnetic enlightment in bacteria. So, are we satisfied with our BRET design? The answer is no; the BRET method is good but not good enough in our project. | The advantage of BRET is that it is a non-radioactive and homogeneous technology. There is no auto-fluorescence coming from compounds or cell and buffer components as no light source is required. And the most of all, it make the conformation change of Mms13 possible to induce optomagnetic enlightment in bacteria. So, are we satisfied with our BRET design? The answer is no; the BRET method is good but not good enough in our project. | ||
| - | + | ==The BiFC-based BRET Phenomenon== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | |
It is known that transmembrane (TM) protein usually have high propensities between the inter-helices. The fact that Mms13 has two tightly interacted helices in transmembrane domain is a big obstacle in our optomagnetic design because the light induced with BRET will keep lightening and cause the neurons excited or inhibited all the time. However, the perfect model we propose is to have the wobbling light, which can function as both ‘’on and off’’ switch for neurons. | It is known that transmembrane (TM) protein usually have high propensities between the inter-helices. The fact that Mms13 has two tightly interacted helices in transmembrane domain is a big obstacle in our optomagnetic design because the light induced with BRET will keep lightening and cause the neurons excited or inhibited all the time. However, the perfect model we propose is to have the wobbling light, which can function as both ‘’on and off’’ switch for neurons. | ||
| Line 43: | Line 31: | ||
In our BiFC-based BRET design, we anchored a half of YFP fragment (with YFP C-terminus) on the inter-helical inhibitor, which also named as the CHAMP peptide design in the following paragraph. By using the inhibitor, we can affect the tight interaction between Mms13’s two helices, and the constant light induced form BRET phenomenon can also be blocked. The rest of the design is to get the other half of YFP (with YFP N-terminus) anchored on the N-terminus of Mms13 and anchored the r-Luciferase on C-terminus of Mms13. The result of our project now will be when there is no magnetic force applied to Mms13, two helices of Mms13 will have farther inter-helical distance due to the inhibitor, CHAMP design and the BRET phenomenon will not perform because the lack of close proximity between two helices. However, when we apply magnetic force to the Mms13, the helices of transmembrane protein will be pulled together and induce the BRET phenomenon to emit fluorescence. | In our BiFC-based BRET design, we anchored a half of YFP fragment (with YFP C-terminus) on the inter-helical inhibitor, which also named as the CHAMP peptide design in the following paragraph. By using the inhibitor, we can affect the tight interaction between Mms13’s two helices, and the constant light induced form BRET phenomenon can also be blocked. The rest of the design is to get the other half of YFP (with YFP N-terminus) anchored on the N-terminus of Mms13 and anchored the r-Luciferase on C-terminus of Mms13. The result of our project now will be when there is no magnetic force applied to Mms13, two helices of Mms13 will have farther inter-helical distance due to the inhibitor, CHAMP design and the BRET phenomenon will not perform because the lack of close proximity between two helices. However, when we apply magnetic force to the Mms13, the helices of transmembrane protein will be pulled together and induce the BRET phenomenon to emit fluorescence. | ||
| - | + | ==The CHAMP Design== | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | == | + | |
CHAMP, the computed helical anti-membrane protein, is one of the computational and genetic methods available to engineer antibody-like molecules that target the water-soluble regions of tansmembrane (TM) proteins. (Hang Yin, et al., 2007) | CHAMP, the computed helical anti-membrane protein, is one of the computational and genetic methods available to engineer antibody-like molecules that target the water-soluble regions of tansmembrane (TM) proteins. (Hang Yin, et al., 2007) | ||
| Line 54: | Line 37: | ||
In the project, we want to use this designed peptide, CHAMP sequence, to inhibit the tight interaction between two helices of membrane protein Mms13 so that we can successfully use mechanical force to change the conformation of Mms13 to induce the BiFC-based BRET phenomenon. The detail part of the CHAMP design is shown in [[link here|protein modelling]]. | In the project, we want to use this designed peptide, CHAMP sequence, to inhibit the tight interaction between two helices of membrane protein Mms13 so that we can successfully use mechanical force to change the conformation of Mms13 to induce the BiFC-based BRET phenomenon. The detail part of the CHAMP design is shown in [[link here|protein modelling]]. | ||
| + | ==Our Constructs== | ||
| + | We have six constructs for our design including both positive and negative controls. | ||
| + | [[Image: c1.png|frame|Fig. 3: The main design construct. Ori is the origin of replication of AMB-1. Pmsp1 is the promoter for AMB-1.]] | ||
| - | + | ==The Modeling of Optomagnetic Design== | |
| - | + | Though we have already done the design part of our project, we may be challenged by whether it can function in reality or not. We consulted with Professor Chun-Min Lo who have professional background in biomechanics. One of his researches is to apply magnetic force on the super magnetic particles contact on the cell membrane and measured the cell’s morphology and biomechanics to study the growth of cells. After we told him the project and our final goal to achieve, he showed the interests and was willing to help us in calculating and modeling for our optomagnetic design and he also generously applied electromagnets, a type of magnet in which the magnetic field is produced by the flow of electric current, in his lab. We then model and calculate the force and magnetic field applied to the magnetic bacteria by using the Professor Chun-Min Lo’s devices. (CM Lo, et al., 1998) | |
| - | + | [[Image: fdevice.png|frame|Fig. 4: Magnetic bead force application. A schematic of the experimental setup, with the electromagnet device shown in cross section. (Joseph N. Fass, et al., 2003) The differences between our project and the paper are that there is no need to bind the magnetic particles on cell membrane, since AMB-1 has its own magnetic beads, magnetosomes, and AMB-1 can be simply put on the medium they used to live and grow without adding other chemicals.]] | |
| + | ===Magnetic force calculations=== | ||
| + | We calculated the force per unit volume of magnetosome by electronic balance, and also from the measured field of the permanent magnet. (Glogauer and Ferrier, 1998) The magnitude of the magnetic force per unit volume of iron oxide beads would be | ||
| - | [[Image: | + | [[Image: ff.png]] |
| - | + | Where M is the magnetization of the beads and δBz/δz is the gradient of the magnetic field along the normal line to the center of the pole face. | |
| - | + | The magnetosome crystal is known to have high chemical purity so its magnetization can reach to the saturation limit for 3*10^5 A/m. The average diameter of the magnetosome is 46 nm in magnetic bacteria, so the volume per bead will be 4.077*10^(-22) m^3. By calculating the data in the paper, we discovered that the cell can have very large scale of deformation when only around 2 nN is applied. Just 2 nN force can that cell We then trace back to the gradient of the magnetic field we should apply to the bacteria. | |
| - | + | The average number of magnetosomes in magnetic bacteria is 20 prticles. So we should have 2 nN/20=0.1 nN applied to one magnetosome. Divided by the magnetosome volume(4.077*10^(-22) m^3), there would be 0.1 nN / (4.077*10^(-22) m^3)≈2.453*10^11 N/m^3 for force per volume. Then, the gradient we need can be counted by dividing the force applied per volume to the saturated magnetization, and it will be (2.453*10^11 N/m^3)/(3*10^5 A/m)≈8.176*10^5 T/m. | |
| + | However, we certainly do not need the high gradient up to 8.176*10^5 T/m. Because the need to pull the membrane and change the conformation of proteins would not have to apply that much force. If the devices we apply now would not strong enough to change Mms13’s conformation. There is still nothing to worry about because we can add more turns per unit length of electromagnet to increase the magnetic gradient and thus induce stronger force to fulfill our goal. | ||
| + | [[Image: electromagnet.png|frame|Fig. 5: Neurite initiation and elongation in response to applied force for only 450 pN. (Joseph N. Fass, et al., 2003)]] | ||
| + | [[Image: electromagnet.png|frame|Fig. 6: A magnetic trap for high-force cell probing is typically composed of an electromagnet with a sharp tip on a micromanipulator. (Hayden Huang, et al., 2004) If the force is not strong enough to change Mms13’s conformation we can add more turns per unit length of electromagnet to increase the magnetic gradient and thus induce stronger force to fulfill our goal.]] | ||
| + | [[Image: device.png|frame|Fig. 7: Measurements of magnetic devices-Electromagnet Gausses at Measured Distances by AZeer Enterprises, LLC]] | ||
| - | + | ==References== | |
| - | + | ||
| - | + | ||
| - | == | + | |
* Johan Bacart, Caroline Corbel, Ralf Jockers, Stéphane Bach, Cyril Couturier Dr. (2008) The BRET technology and its application to screening assays. Biotechnology Journal Volume 3, Issue 3: 311–324 | * Johan Bacart, Caroline Corbel, Ralf Jockers, Stéphane Bach, Cyril Couturier Dr. (2008) The BRET technology and its application to screening assays. Biotechnology Journal Volume 3, Issue 3: 311–324 | ||
| Line 92: | Line 81: | ||
DeGrado (2007). Supporting Online Material for Computational Design of Peptides That Target Transmembrane Helices. Science 315, 1817. | DeGrado (2007). Supporting Online Material for Computational Design of Peptides That Target Transmembrane Helices. Science 315, 1817. | ||
| - | *J. S. Slusky, H. Yin and W. F. DeGrado (2009) | + | * J. S. Slusky, H. Yin and W. F. DeGrado. (2009) Understanding Membrane Proteins. How to Design Inhibitors of Transmembrane Protein—Protein Interactions. PROTEIN ENGINEERING Nucleic Acids and Molecular Biology 22, 315-337. |
| + | |||
| + | * Michael Glogauer and J. Ferrier. (1998) A new method for application of force to cells via ferric oxide beads. PFLÜGERS ARCHIV EUROPEAN JOURNAL OF PHYSIOLOGY 435, 2:320-327. | ||
| + | |||
| + | * Chun-Min Lo, Michael Glogauer, Marisa Rossi and J. Ferrier. (1998) Cell-substrate separation: effect of applied force and temperature. EUROPEAN BIOPHYSICS JOURNAL 27, 1: 9-17 | ||
| + | |||
| + | * Hayden Huang, Roger D. Kamm, and Richard T. Lee. (2004) Cell mechanics and mechanotransduction: pathways, probes, and physiology. Am J Physiol Cell Physiol 287: C1–C11. | ||
| + | |||
| + | * Joseph N. Fass and David J. Odde. (2003) Tensile Force-Dependent Neurite Elicitation via Anti-b1 Integrin Antibody-Coated Magnetic Beads. Biophysical Journal Volume 85:623–636 623. | ||
| + | |||
| + | * Hayden Huang, Jeremy Sylvan, Maxine Jonas, Rita Barresi, Peter T. C. So, Kevin P. Campbell, and Richard T. Lee. (2005) Cell stiffness and receptors: evidence for cytoskeletal subnetworks. Am J Physiol Cell Physiol 288: C72–C80. | ||
Revision as of 11:49, 5 October 2011

==Optomagnetic Design==(by 尚叡)
==Magnetic Bacteria==(by 尚叡)
==The Anchor Protein==(by 尚叡)
Contents |
The BRET Phenomenon
How to use the conformation change of the Mms13 transmembrane protein to induce the light? We first came up with the idea by using bioluminescence resonance energy transfer (BRET). BRET is based on resonance energy transfer between a light-emitting enzyme and a fluorescent acceptor. (Johan Bacart, et al., 2008) For example, the energy derived from a luciferase reaction can be used to excite a fluorescent protein when the latter is in close proximity to the luciferase enzyme. Several other methods developed are shown in Table 1.
We chose the original BRET method using coelenterazine (luciferin) as substrate in BRET 1 method (See Figure 1). What we do is anchoring the YFP on the N-terminus of Mms13 and r-Luciferase on the C-terminus. We expect that when the magnetic force applied on the membrane and changed the conformation of Mms13 would lead to the close proximity between YFP and r-Luciferase and thus induce fluorescent phenomenon by BRET method (See Figure 1).
The advantage of BRET is that it is a non-radioactive and homogeneous technology. There is no auto-fluorescence coming from compounds or cell and buffer components as no light source is required. And the most of all, it make the conformation change of Mms13 possible to induce optomagnetic enlightment in bacteria. So, are we satisfied with our BRET design? The answer is no; the BRET method is good but not good enough in our project.
The BiFC-based BRET Phenomenon
It is known that transmembrane (TM) protein usually have high propensities between the inter-helices. The fact that Mms13 has two tightly interacted helices in transmembrane domain is a big obstacle in our optomagnetic design because the light induced with BRET will keep lightening and cause the neurons excited or inhibited all the time. However, the perfect model we propose is to have the wobbling light, which can function as both ‘’on and off’’ switch for neurons.
Luckily, we found the inspiration from one paper associated with the bimolecular fluorescence complementation (BiFC) method. The BiFC assay is based on the association between two nonfluorescent fragments of a fluorescent protein. When those fragments are brought in proximity to each other by an interaction between proteins, they would fuse into one functional fluorescent protein. (Tom K. Kerppola., 2008) We came up with the idea by mixing the BiFC method and BRET phenomenon into the BiFC-based BRET design. (See Figure 2)
In our BiFC-based BRET design, we anchored a half of YFP fragment (with YFP C-terminus) on the inter-helical inhibitor, which also named as the CHAMP peptide design in the following paragraph. By using the inhibitor, we can affect the tight interaction between Mms13’s two helices, and the constant light induced form BRET phenomenon can also be blocked. The rest of the design is to get the other half of YFP (with YFP N-terminus) anchored on the N-terminus of Mms13 and anchored the r-Luciferase on C-terminus of Mms13. The result of our project now will be when there is no magnetic force applied to Mms13, two helices of Mms13 will have farther inter-helical distance due to the inhibitor, CHAMP design and the BRET phenomenon will not perform because the lack of close proximity between two helices. However, when we apply magnetic force to the Mms13, the helices of transmembrane protein will be pulled together and induce the BRET phenomenon to emit fluorescence.
The CHAMP Design
CHAMP, the computed helical anti-membrane protein, is one of the computational and genetic methods available to engineer antibody-like molecules that target the water-soluble regions of tansmembrane (TM) proteins. (Hang Yin, et al., 2007)
In the project, we want to use this designed peptide, CHAMP sequence, to inhibit the tight interaction between two helices of membrane protein Mms13 so that we can successfully use mechanical force to change the conformation of Mms13 to induce the BiFC-based BRET phenomenon. The detail part of the CHAMP design is shown in protein modelling.
Our Constructs
We have six constructs for our design including both positive and negative controls.
The Modeling of Optomagnetic Design
Though we have already done the design part of our project, we may be challenged by whether it can function in reality or not. We consulted with Professor Chun-Min Lo who have professional background in biomechanics. One of his researches is to apply magnetic force on the super magnetic particles contact on the cell membrane and measured the cell’s morphology and biomechanics to study the growth of cells. After we told him the project and our final goal to achieve, he showed the interests and was willing to help us in calculating and modeling for our optomagnetic design and he also generously applied electromagnets, a type of magnet in which the magnetic field is produced by the flow of electric current, in his lab. We then model and calculate the force and magnetic field applied to the magnetic bacteria by using the Professor Chun-Min Lo’s devices. (CM Lo, et al., 1998)

Magnetic force calculations
We calculated the force per unit volume of magnetosome by electronic balance, and also from the measured field of the permanent magnet. (Glogauer and Ferrier, 1998) The magnitude of the magnetic force per unit volume of iron oxide beads would be
Where M is the magnetization of the beads and δBz/δz is the gradient of the magnetic field along the normal line to the center of the pole face.
The magnetosome crystal is known to have high chemical purity so its magnetization can reach to the saturation limit for 3*10^5 A/m. The average diameter of the magnetosome is 46 nm in magnetic bacteria, so the volume per bead will be 4.077*10^(-22) m^3. By calculating the data in the paper, we discovered that the cell can have very large scale of deformation when only around 2 nN is applied. Just 2 nN force can that cell We then trace back to the gradient of the magnetic field we should apply to the bacteria.
The average number of magnetosomes in magnetic bacteria is 20 prticles. So we should have 2 nN/20=0.1 nN applied to one magnetosome. Divided by the magnetosome volume(4.077*10^(-22) m^3), there would be 0.1 nN / (4.077*10^(-22) m^3)≈2.453*10^11 N/m^3 for force per volume. Then, the gradient we need can be counted by dividing the force applied per volume to the saturated magnetization, and it will be (2.453*10^11 N/m^3)/(3*10^5 A/m)≈8.176*10^5 T/m.
However, we certainly do not need the high gradient up to 8.176*10^5 T/m. Because the need to pull the membrane and change the conformation of proteins would not have to apply that much force. If the devices we apply now would not strong enough to change Mms13’s conformation. There is still nothing to worry about because we can add more turns per unit length of electromagnet to increase the magnetic gradient and thus induce stronger force to fulfill our goal.

References
- Johan Bacart, Caroline Corbel, Ralf Jockers, Stéphane Bach, Cyril Couturier Dr. (2008) The BRET technology and its application to screening assays. Biotechnology Journal Volume 3, Issue 3: 311–324
- Kevin D G Pfleger, Karin A Eidne. (2006) Illuminating insights into protein-protein interactions using bioluminescence resonance energy transfer (BRET). Nature Methods 3:165 - 174
- Tom K. Kerppola. (2008) Bimolecular Fluorescence Complementation (BiFC) Analysis as a Probe of Protein Interactions in Living Cells. Annu Rev Biophys 37: 465–487.
- Hang Yin, Joanna S. Slusky, Bryan W. Berger, Robin S. Walters, Gaston Vilaire,
Rustem I. Litvinov, James D. Lear, Gregory A. Caputo, Joel S. Bennett, William F. DeGrado (2007). Supporting Online Material for Computational Design of Peptides That Target Transmembrane Helices. Science 315, 1817.
- J. S. Slusky, H. Yin and W. F. DeGrado. (2009) Understanding Membrane Proteins. How to Design Inhibitors of Transmembrane Protein—Protein Interactions. PROTEIN ENGINEERING Nucleic Acids and Molecular Biology 22, 315-337.
- Michael Glogauer and J. Ferrier. (1998) A new method for application of force to cells via ferric oxide beads. PFLÜGERS ARCHIV EUROPEAN JOURNAL OF PHYSIOLOGY 435, 2:320-327.
- Chun-Min Lo, Michael Glogauer, Marisa Rossi and J. Ferrier. (1998) Cell-substrate separation: effect of applied force and temperature. EUROPEAN BIOPHYSICS JOURNAL 27, 1: 9-17
- Hayden Huang, Roger D. Kamm, and Richard T. Lee. (2004) Cell mechanics and mechanotransduction: pathways, probes, and physiology. Am J Physiol Cell Physiol 287: C1–C11.
- Joseph N. Fass and David J. Odde. (2003) Tensile Force-Dependent Neurite Elicitation via Anti-b1 Integrin Antibody-Coated Magnetic Beads. Biophysical Journal Volume 85:623–636 623.
- Hayden Huang, Jeremy Sylvan, Maxine Jonas, Rita Barresi, Peter T. C. So, Kevin P. Campbell, and Richard T. Lee. (2005) Cell stiffness and receptors: evidence for cytoskeletal subnetworks. Am J Physiol Cell Physiol 288: C72–C80.
 "
"