Team:HKU-Hong Kong/Lab Diaries
From 2011.igem.org
(Difference between revisions)
| Line 5: | Line 5: | ||
|style="width:900px;"|'''Week 1''' | |style="width:900px;"|'''Week 1''' | ||
Transformation of reporter DNA (pEGFP-loxp-km-loxp) into DH10B (non-virulent strain E. coli) with antibiotic resistance (Chloramphenicol – Cm) | Transformation of reporter DNA (pEGFP-loxp-km-loxp) into DH10B (non-virulent strain E. coli) with antibiotic resistance (Chloramphenicol – Cm) | ||
| - | |||
|- | |- | ||
|style="width:900px;"|'''Week 2''' | |style="width:900px;"|'''Week 2''' | ||
| Line 24: | Line 23: | ||
<LI>2uL of 10-fold diluted pEGFP-loxp-km-loxp- tetO2 – 3 was transformed into DH10B to greatly amplify the product by using bacterial cells. | <LI>2uL of 10-fold diluted pEGFP-loxp-km-loxp- tetO2 – 3 was transformed into DH10B to greatly amplify the product by using bacterial cells. | ||
</OL> | </OL> | ||
| - | |||
|- | |- | ||
|style="width:900px;"|'''Week 3''' | |style="width:900px;"|'''Week 3''' | ||
| Line 73: | Line 71: | ||
<LI>Reverse PCR was used to insert tetO2 -0 and tetO2 – 4 into pEGFP-loxp-km-loxp. | <LI>Reverse PCR was used to insert tetO2 -0 and tetO2 – 4 into pEGFP-loxp-km-loxp. | ||
<LI>2uL of pEGFP-loxp-km-loxp- tetO2 – 0 and pEGFP-loxp-km-loxp- tetO2 – 4 were transformed into DH10B separately to greatly amplify the product by using bacterial cells. | <LI>2uL of pEGFP-loxp-km-loxp- tetO2 – 0 and pEGFP-loxp-km-loxp- tetO2 – 4 were transformed into DH10B separately to greatly amplify the product by using bacterial cells. | ||
| - | |||
|- | |- | ||
|style="width:900px;"|'''Week 6''' | |style="width:900px;"|'''Week 6''' | ||
| Line 92: | Line 89: | ||
</OL> | </OL> | ||
</OL> | </OL> | ||
| - | |||
|- | |- | ||
|style="width:900px;"|'''Week 7''' | |style="width:900px;"|'''Week 7''' | ||
| Line 132: | Line 128: | ||
|} | |} | ||
</div> | </div> | ||
| - | |||
|- | |- | ||
|style="width:900px;"|'''Week 10''' | |style="width:900px;"|'''Week 10''' | ||
| Line 151: | Line 146: | ||
<LI>Fluorescence intensity were checked at OD600 using spectrophotometer | <LI>Fluorescence intensity were checked at OD600 using spectrophotometer | ||
</OL> | </OL> | ||
| - | |||
|- | |- | ||
|style="width:900px;"|'''Week 11''' | |style="width:900px;"|'''Week 11''' | ||
| Line 191: | Line 185: | ||
</OL> | </OL> | ||
<LI>Purification of NdeI digested pET28a | <LI>Purification of NdeI digested pET28a | ||
| - | |||
|- | |- | ||
|style="width:900px;"|'''Week 12''' | |style="width:900px;"|'''Week 12''' | ||
| Line 207: | Line 200: | ||
<LI>tetO2 – 1 | <LI>tetO2 – 1 | ||
<LI>sfGFP | <LI>sfGFP | ||
| - | |||
|- | |- | ||
|style="width:900px;"|'''Week 13''' | |style="width:900px;"|'''Week 13''' | ||
| Line 228: | Line 220: | ||
<LI>J23103RH | <LI>J23103RH | ||
<LI>J23116R | <LI>J23116R | ||
| - | |||
|- | |- | ||
|style="width:900px;"|'''Week 14''' | |style="width:900px;"|'''Week 14''' | ||
Revision as of 06:57, 5 October 2011
| Lab Diaries | |
| Week 1
Transformation of reporter DNA (pEGFP-loxp-km-loxp) into DH10B (non-virulent strain E. coli) with antibiotic resistance (Chloramphenicol – Cm) | |
Week 2
| |
Week 3
| Lab Diaries |
| Week 4-5
tetO2 – 0 and tetO2 – 4
| |
| Week 6
Overlap PCR was carried out to produce fusion protein gene
| |
| Week 7
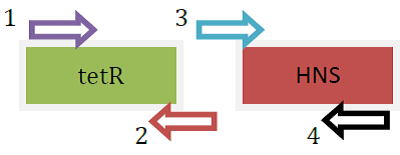
Overlap PCR was carried out to produce fusion protein gene tetR::HNS (2-90)
| Lab Diaries |
Week 8
| Lab Diaries |
Week 9
| |
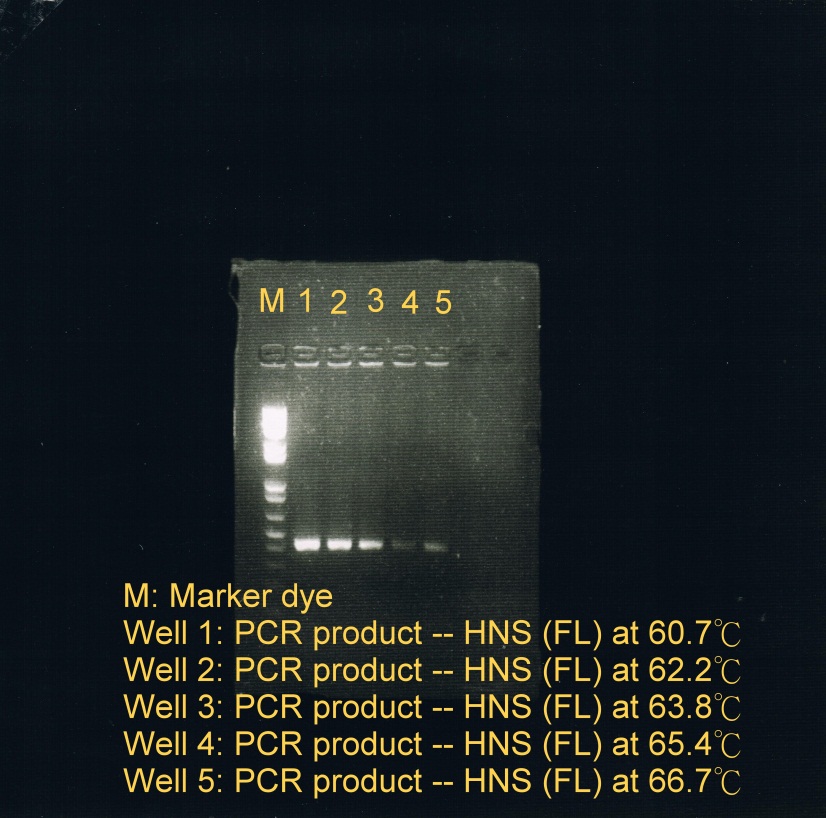
Week 10
| |
Week 11
| |
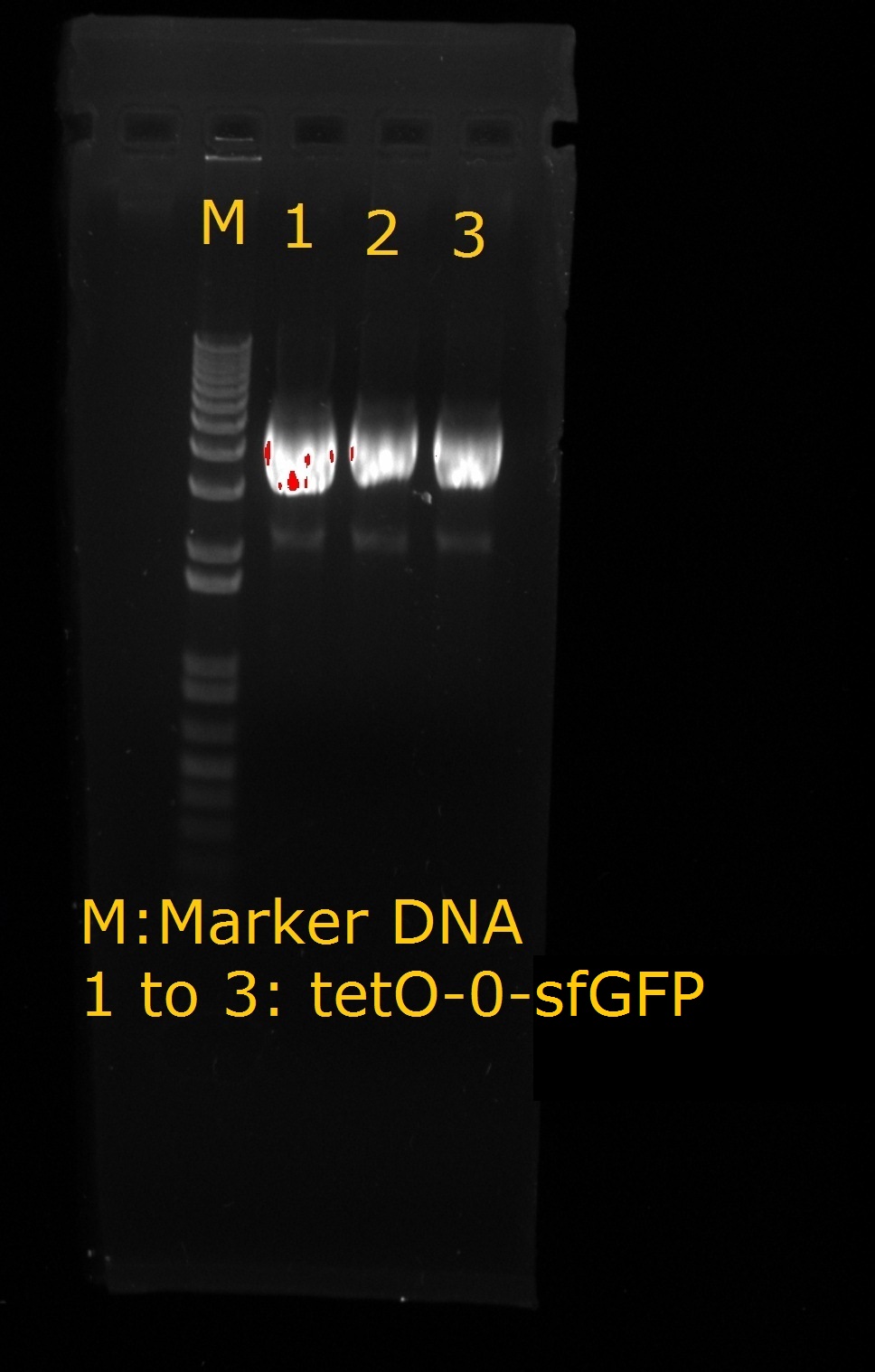
Week 12
| |
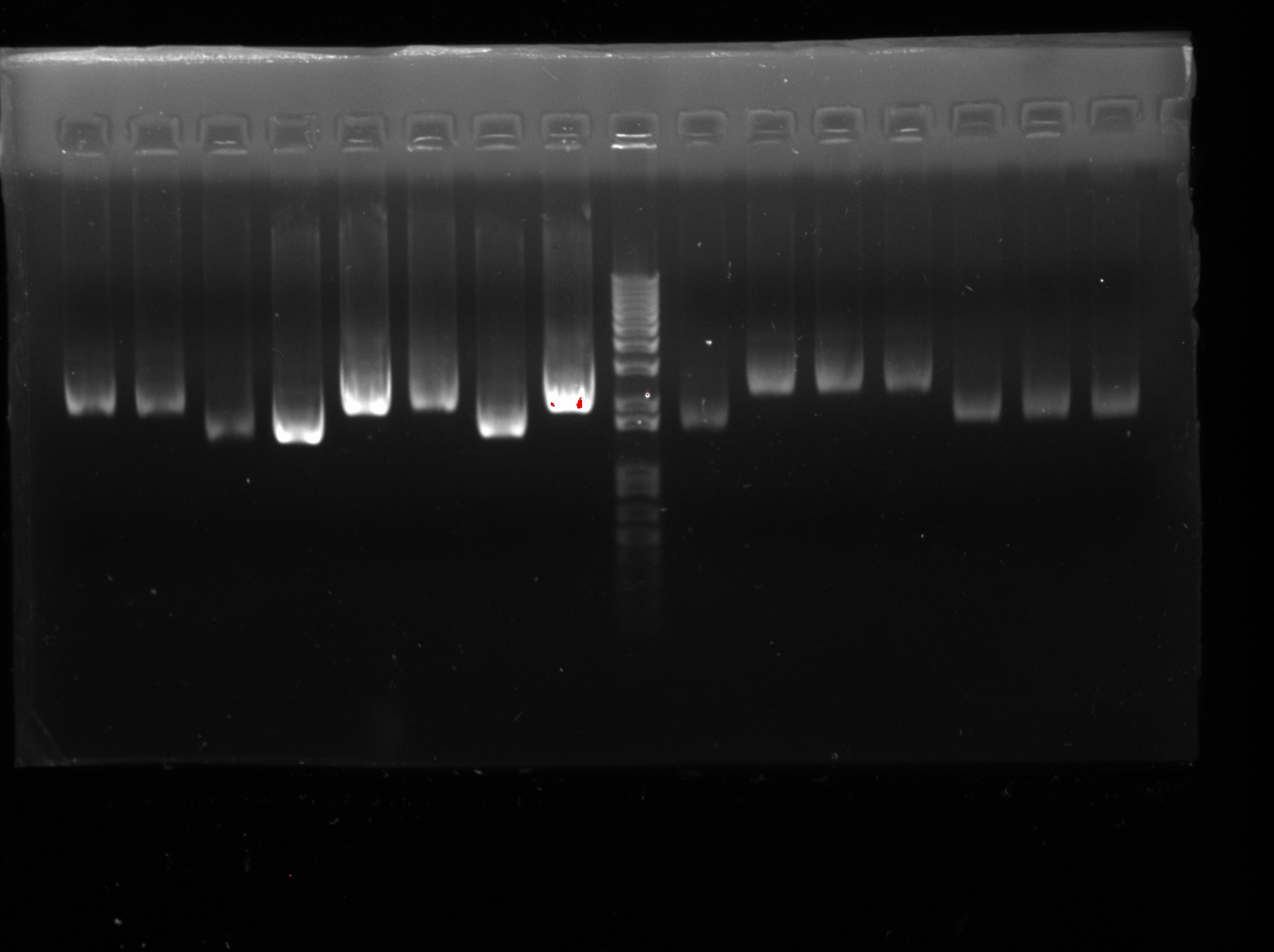
Week 13
| |
Week 14
|
 "
"