From 2011.igem.org
(Difference between revisions)
|
|
| Line 59: |
Line 59: |
| | |} | | |} |
| | </div> | | </div> |
| | + | |style="font-family: georgia, helvetica, arial, sans-serif;font-size:2em;color:#01DF01;"|Lab Diaries |
| | + | |- |
| | + | |style="width:900px;"|'''Week 4-5''' |
| | + | <OL> |
| | + | <LI>tetO2 – 0 and tetO2 – 4 |
| | + | <OL> |
| | + | <LI>Reverse PCR was used to insert tetO2 -0 and tetO2 – 4 into pEGFP-loxp-km-loxp |
| | + | <LI>2uL of pEGFP-loxp-km-loxp- tetO2 – 0 and pEGFP-loxp-km-loxp- tetO2 – 4 were transformed into DH10B separately to greatly amplify the product by using bacterial cells |
Revision as of 17:17, 4 October 2011
| Lab Diaries
|
| Week 1
Transformation of reporter DNA (pEGFP-loxp-km-loxp) into DH10B (non-virulent strain E. coli) with antibiotic resistance (Chloramphenicol – Cm)
| Lab Diaries
|
Week 2
- tetO2 -1 (DNA binding site)
- Reverse PCR was used to insert tetO2 -1 into pEGFP-loxp-km-loxp (template DNA) by using forward and reverse primers which are with the tetO2 -1. By using reverse PCR, we can insert the tetO2 -1 site into the pEGFP while the plasmid produced is still in double-stranded circular form.
- pEGFP-loxp-km-loxp-tetO2-1 was then transformed into DH10B to greatly amplify the product by using bacterial cells
- tetO2 -2 (DNA biding site)
- Reverse PCR was used to insert tetO2 -2 into pEGFP-loxp-km-loxp
- 2uL of 10-fold diluted pEGFP-loxp-km-loxp- tetO2 -2 was transformed into DH10B to greatly amplify the product by using bacterial cells
- tetO2 – 3 (DNA binding site)
- Reverse PCR was used to insert tetO2 – 3 into pEGFP-loxp-km-loxp
- 2uL of 10-fold diluted pEGFP-loxp-km-loxp- tetO2 – 3 was transformed into DH10B to greatly amplify the product by using bacterial cells
| Lab Diaries
|
Week 3
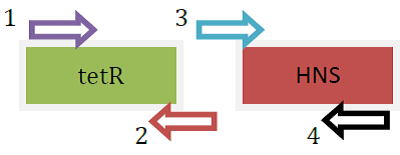
- Overlap PCR (involving 3 steps) was carried out to produce the fusion protein [tetR-HNS (any length)]
- PCR was carried out separately for 2 genes, tetR and HNS (full length, in this case)
- Primers used were as below:
- tetR: [forward] R-H-out-F; [reverse] R-H-tetR-R
- HNS (FL): [forward] R-H-tetR-F-out; [reverse] R-H(FL)-2-R
|
| Fusion protein genes produced separately using PCR
|
- Another PCR was carried out using primers with linker to insert the linker DNA to the tetR and HNS (FL) for the fusion of the 2 genes.
- PCR was used to further amplify the product from step 2 (fusion protein gene)
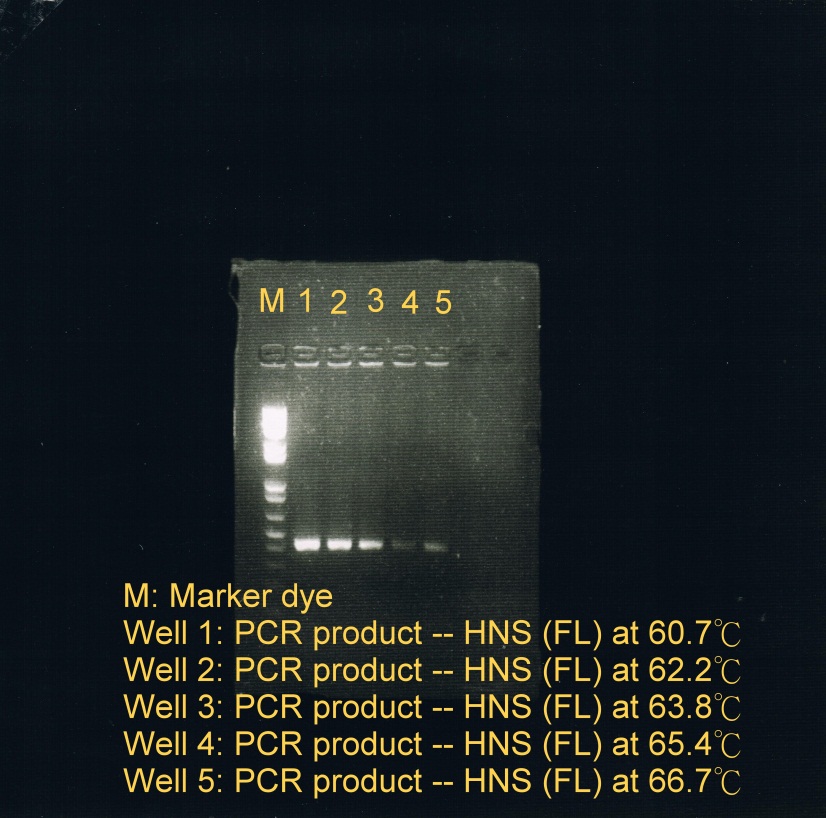
- PCR was used to obtain HNS(FL) at 5 different temperatures to test which temperature is optimal for annealing
- Annealing phase at 5 different temperatures: (a) 60.7C; (b) 62.2C; (c) 63.8C; (d) 65.4C; (e) 66.7C (less sample)
- Electrophoresis was carried to determine which temperature best suit the annealing phase. From the gel image, it was observed that a high concentration of products was resulted at annealing temperature between 60.7C and 62.2C.
|
| Fusion protein genes annealed at 5 different temperatures
|
| Lab Diaries
|
Week 4-5
- tetO2 – 0 and tetO2 – 4
- Reverse PCR was used to insert tetO2 -0 and tetO2 – 4 into pEGFP-loxp-km-loxp
- 2uL of pEGFP-loxp-km-loxp- tetO2 – 0 and pEGFP-loxp-km-loxp- tetO2 – 4 were transformed into DH10B separately to greatly amplify the product by using bacterial cells
|
 "
"




