Team:Tsinghua/experiment
From 2011.igem.org
(→Molecular cloning) |
(→Expression Test) |
||
| Line 39: | Line 39: | ||
| colspan="2" | 3ml | | colspan="2" | 3ml | ||
|- | |- | ||
| - | | IPTG(mM) | + | | rowspan="2" | IPTG(mM) |
| colspan="2" | 0.1 | | colspan="2" | 0.1 | ||
|- | |- | ||
Revision as of 14:09, 4 October 2011

Present proteins to the outer membrane
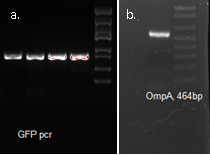
We used OmpA-His tag-GFP to test the conditions for presenting protein to the outer membrane. The design allows double check whether the C-terminal moeity can be presented outside. One is by green fluorescence, the other is by Western blotting with antibody against his tag.
Molecular cloning
OmpA, GFP His tag 1507bp confirmed by sequencing
Expression Test
Test for proper inducing conditions
| Group | 1 | 2 |
|---|---|---|
| Volume | 3ml | |
| IPTG(mM) | 0.1 | |
| 0.5 | ||
| 1.0 | ||
| Temperature | 30oC | 18oC |
| Duration | 12h | 12h |
| Result | Media remained yellowish. E. coli white | Media turned greenish. E. coli green |
Use 0.5mM IPTG, 18 centigrade, 12 hours induction in later experiments.
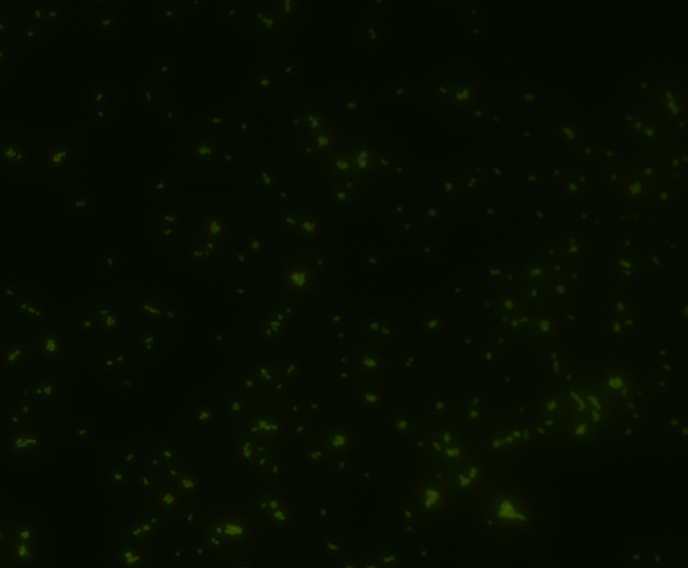
Fluorescence microscopy
Green fluorescence is bright.
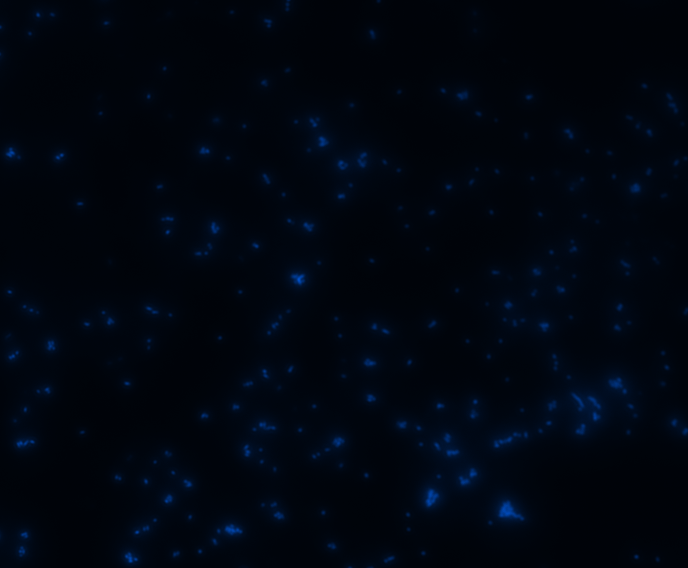
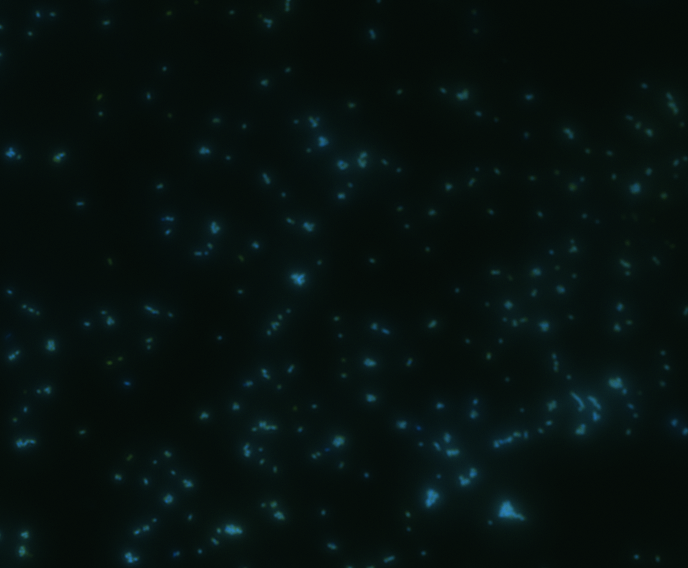
In order to determine whether GFP is presented outside the membrane, cells were digested with Protease K at 50 centigrade for 30min.
| Color Channel | Pre-digest | Post-digest |
|---|---|---|
| GFP | 
| 
|
| DAPI | 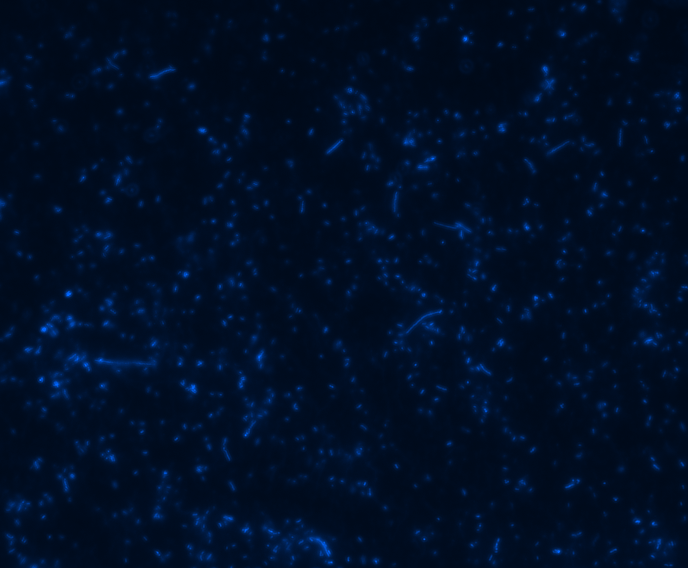
| 
|
| Merge | 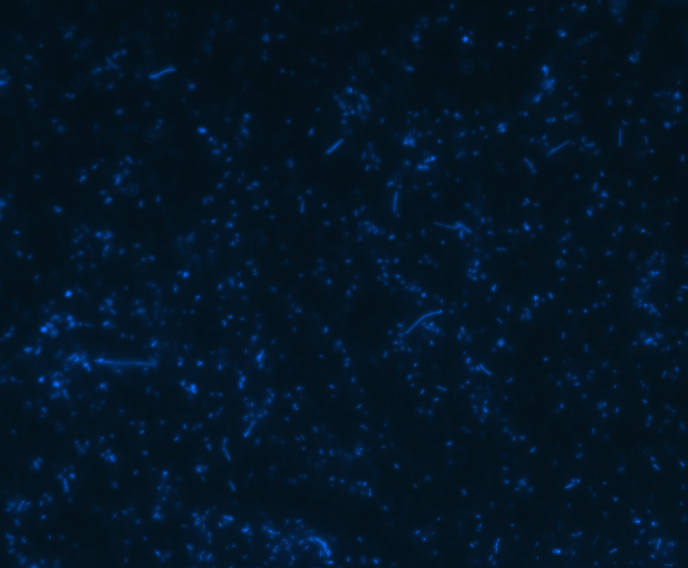
| 
|
The fluorescence disappeared completely after digestion, which seems too good to be true. Hence, we performed Western blotting to be sure of the result.
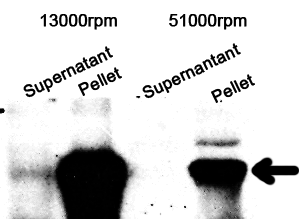
Western Blotting
The cell components were first fractionated before blotting.
1. Sonicate the bacteria culture.
2. Centrifuge 13000rpm, 30min to separate soluble protein and membrane components from the cell debris and the non-soluble protein.
3. 51000rpm 1h to separate soluble protein from membrane components.
The majority of the OmpA-GFP protein is in the pellet after the first centrifuge, indicating incomplete cell lysis or inefficient protein folding. Nonetheless, from the later result, we can see that it is exclusively in the membrane and not in the soluble proteins.
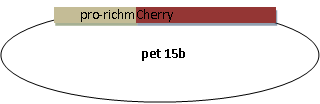
Expression of Substrate
Molecular cloning
We synthesized the multi-proline sequence and linked it to mCherry.
Test of Expression
| Group | 1 | 2 |
|---|---|---|
| Volume | 3ml | |
| IPTG(mM) | 0.1 | |
| 0.5 | ||
| 1.0 | ||
| Temperature | 30oC | 18oC |
| Duration | 12h | 12h |
| Result | Media remained yellowish. E. coli white | Media turned greenish. E. coli green |
2. pet15b OmpA-HIV cleavage site-sh3 OmpA-sh3 (920bp), add HIV cleavage site to 968bp. Confirmed by sequencing
3. pet15b pro-rich –mCherry confirmed by sequencing
4. pet15b OmpA-HIV protease
1045bp confirmed by sequencing
2. Protein expression test 1. Pro rich-mCherry a. Test for proper inducing condition(table2) Table 2 group 1 2 Volume 3ml IPTG(mM) 0 0.1 0.5 1 Temperature 30℃ 18℃ Duration 5h 13h description Medium remained yellow. E.coli white Medium turned red E.coli cherry pink
After Pro-rich mCherry expression, significant color change can be observed (bacteria cells turned cherry red as a result of mCherry expression)
1-a b. SDSPAGE(Coomassie brilliant blue stain) No significant specific band observed
1 2 3 4 5 6 7 8 Marker 9 10 11 12 13 14
1-b
c. Ni column purification
Elution Description 5mM Flowthrough still red, no color change within column 20mM洗脱 Flowthrough still red 200mM洗脱 Flowthrough red, significant color change within column can be observed(dark red to grey) Coomassie brilliant blue stain after purification
1 2 3 4 5 6 Marker 7 8 9 10
1-c d. Western Blotting(1-d)
图1-d 1. OmpA-GFP membrane transportation a. Test for proper inducing condition(table4) Table 4 Group 1 2 Volumn 3ml IPTG (mM) 0 0.1 0.5 1 Temperature 30℃ 18℃ Duration 7h 12h Description Medium remained yellow. E.coli white Medium slightly changed to green E.coli Green florescence Florescence : green color can be observed by eyes b. Membrane fraction isolation 13000rpm, 30min. Supernatant: soluble protein and membrane fraction Pellet: non-sonicated cell, cell debris, non-soluble protein 51000rpm 1h Supernatant: soluble protein Pellet: Membrane fraction c. Western Blotting
13000rpm 51000rpm
S P S P
 "
"