Team:NYMU-Taipei/synthoprime
From 2011.igem.org
(Difference between revisions)
| Line 16: | Line 16: | ||
| valign=top| | | valign=top| | ||
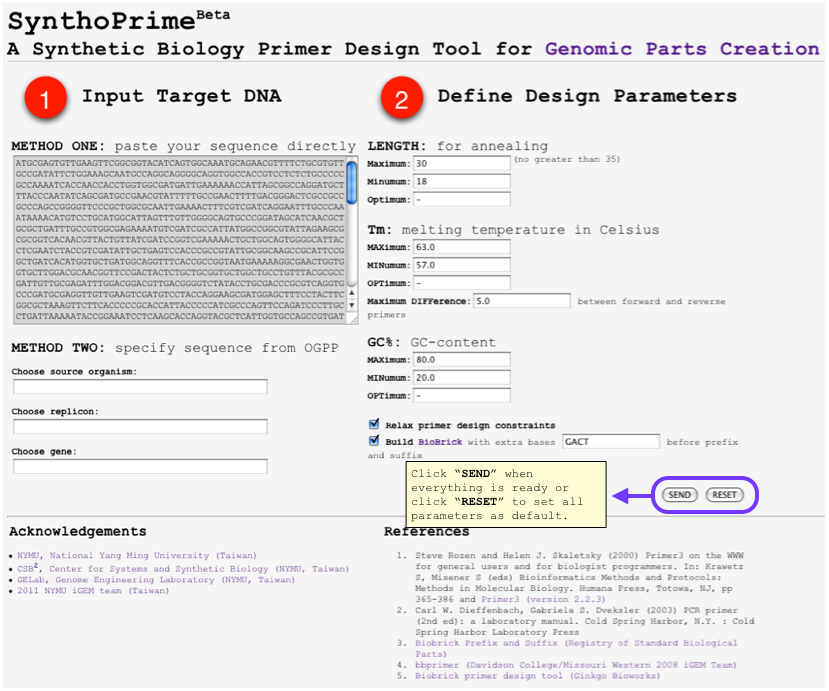
[[File:SynthoPrime manual p4.png|thumb|300px| '''STEP 4:''' Click “SEND” when everything is ready or click “RESET” to set all parameters as default.]] | [[File:SynthoPrime manual p4.png|thumb|300px| '''STEP 4:''' Click “SEND” when everything is ready or click “RESET” to set all parameters as default.]] | ||
| + | |- | ||
| + | |} | ||
| + | |||
| + | HOWTO: Output Reading | ||
| + | |||
| + | {| | ||
| + | | valign=top| | ||
| + | [[File:SynthoPrime manual p5.png|thumb|630px|Main output page]] | ||
| + | |- | ||
| + | | | ||
| + | [[File:SynthoPrime manual p6.png|thumb|630px|Additional Info.]] | ||
|- | |- | ||
|} | |} | ||
Revision as of 09:12, 4 October 2011

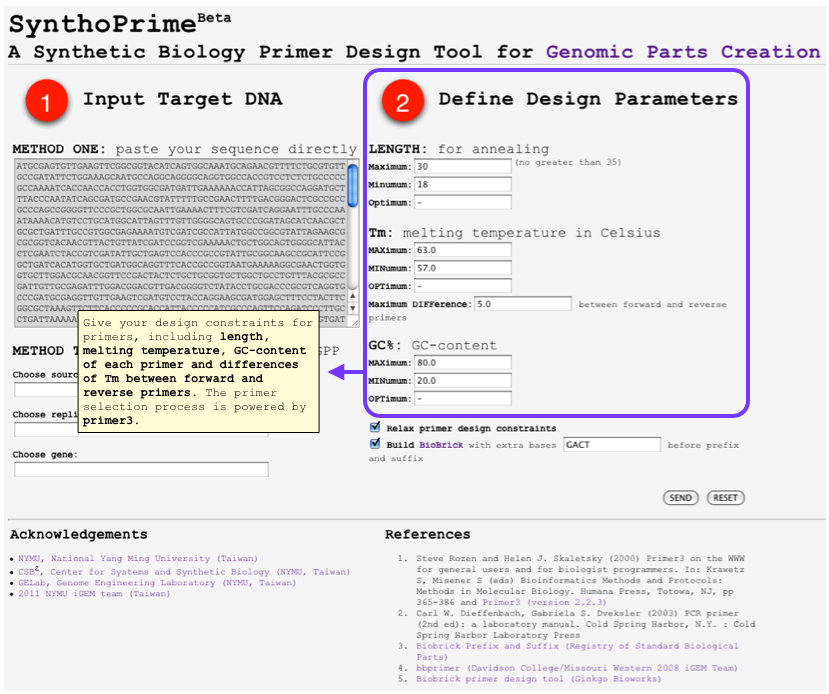
HOWTO: User Inputs
HOWTO: Output Reading
 "
"