Team:Panama/Protocols/AssemblyMethods
From 2011.igem.org
(Difference between revisions)
Ernestopro (Talk | contribs) (→Rational Design) |
Ernestopro (Talk | contribs) (→Rational Design) |
||
| Line 10: | Line 10: | ||
The rationally-egineering for ligate the BioBrick parts was able by using the[http://partsregistry.org/Assembly:3A_Assembly '''3A Assembly method'''] | The rationally-egineering for ligate the BioBrick parts was able by using the[http://partsregistry.org/Assembly:3A_Assembly '''3A Assembly method'''] | ||
| + | |||
| + | |||
| + | [[File:Rational-Design.jpg|up|center|500px]] | ||
| + | |||
| + | |||
[[File:home.jpg|100px|link=https://2011.igem.org/Team:Panama]] | [[File:home.jpg|100px|link=https://2011.igem.org/Team:Panama]] | ||
Revision as of 05:29, 29 September 2011
Assembly Method
How we design and build our BioBrick parts?
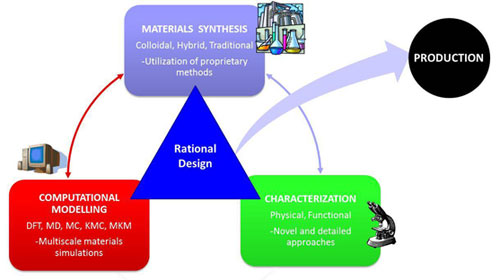
Rational Design
We need to build using the Assembly Standard Protocol 10 an expression platform that works into our modified E.coli. For this, we choose four standardized biological parts from the Registry, those parts are: Promoter T7, Ribosomal Binding Site (RBS), GFP Reporter and Terminator; Were selected using rational design abstraction from molecular biology and genetic engineering to enable us to build up rationally-engineered, our expression platform.
The rationally-egineering for ligate the BioBrick parts was able by using the[http://partsregistry.org/Assembly:3A_Assembly 3A Assembly method]
 "
"