|
|
| (87 intermediate revisions not shown) |
| Line 1: |
Line 1: |
| - | <!-- *** What falls between these lines is the Alert Box! You can remove it from your pages once you have read and understood the alert *** --> | + | <!-- LORAN, this is what comments look like, comments intended for you will begin with "LORAN" --> |
| | | | |
| - | <html> | + | <!-- LORAN, put a better picture here pretty please; it'd be really great if you could make a header which includes the little navigation box and an image, to have at the top of every page (navigation box is currently at the bottom of this page; this random example that I found looks pretty simple, it's just a modified box with an image on top: https://2010.igem.org/Team:Baltimore_US; just press "view source" to see and steal their code --> |
| - | <div id="box" style="width: 700px; margin-left: 137px; padding: 5px; border: 3px solid #000; background-color: #fe2b33;">
| + | <!-- Image at the top of each page --> |
| - | <div id="template" style="text-align: center; font-weight: bold; font-size: large; color: #f6f6f6; padding: 5px;">
| + | <center> |
| - | This is a template page. READ THESE INSTRUCTIONS.
| + | [[Image:division.png|225 px|bottom]] |
| - | </div>
| + | [[Image:Virginia_Tech_logo.png|250px]] |
| - | <div id="instructions" style="text-align: center; font-weight: normal; font-size: small; color: #f6f6f6; padding: 5px;">
| + | [[Image:diatoms.png|210 px|bottom]] |
| - | You are provided with this team page template with which to start the iGEM season. You may choose to personalize it to fit your team but keep the same "look." Or you may choose to take your team wiki to a different level and design your own wiki. You can find some examples <a href="https://2009.igem.org/Help:Template/Examples">HERE</a>.
| + | </center> |
| - | </div> | + | |
| - | <div id="warning" style="text-align: center; font-weight: bold; font-size: small; color: #f6f6f6; padding: 5px;">
| + | |
| - | You <strong>MUST</strong> have a team description page, a project abstract, a complete project description, a lab notebook, and a safety page. PLEASE keep all of your pages within your teams namespace.
| + | |
| - | </div>
| + | |
| - | </div> | + | |
| - | </html>
| + | |
| | | | |
| - | <!-- *** End of the alert box *** --> | + | <!--- Navigation Box ---> |
| | + | <!-- baltimore's box style: "background-color:#7998AD;" cellpadding="1" cellspacing="1" border="0" bordercolor="#fff" width="924px" align="center" --> |
| | + | {| style=" background-color:#800000;" cellpadding="1" cellspacing="5" border="0" width="90%" align="center" |
| | | | |
| | | | |
| | + | !align="center"|[[Team:Virginia_Tech|<span style="color:orange;">Home</span>]] |
| | + | !align="center"|[[Team:Virginia_Tech/Team|<span style="color:orange;">Team</span>]] |
| | + | !align="center"|[https://igem.org/Team.cgi?year=2011&team_name=Virginia_Tech <span style="color:orange;">Official Team Profile</span>] |
| | + | !align="center"|[[Team:Virginia_Tech/Project|<span style="color:orange;">Project</span>]] |
| | + | !align="center"|[[Team:Virginia_Tech/Parts|<span style="color:orange;">Parts Submitted to the Registry</span>]] |
| | + | !align="center"|[[Team:Virginia_Tech/Notebook|<span style="color:orange;">Notebook</span>]] |
| | + | !align="center"|[[Team:Virginia_Tech/Safety|<span style="color:orange;">Safety</span>]] |
| | + | !align="center"|[[Team:Virginia_Tech/Attributions|<span style="color:orange;">Attributions</span>]] |
| | + | |} |
| | | | |
| | + | __NOTOC__ |
| | {|align="justify" | | {|align="justify" |
| - | |<h2>Project Description:</h2>
| + | <h3>Characterization of Fluorescent Reporters:</h3> |
| - | <p>Since the isolation of GFP in 1962, fluorescent proteins have become a ubiquitous tool for studying cellular processes and have recently been used in biosensors. To be effective for this application, however, they will need to be better characterized. To this end, the 2011 VT iGEM team is interested in finding and optimizing good fluorescent reporters which mature and degrade quickly, and which fluoresce brightly. This involves finding fast-folding fluorescent proteins and degradation tags that help destabilize and degrade them. We hope that this work will lay a good foundation for a more quantitative use of fluorescent reporters in the future.</p>
| + | |
| - | <p>To tackle this problem, we have split our team into three specialized groups focusing on the Design, Fabrication, and Characterization of our fluorescent protein constructs. This transdisciplinary approach allows us to work more quickly and achieve more ambitious goals</p>
| + | |
| | | | |
| - | |[[Image:Virginia_Tech_logo.png|100px|right|frame]]
| + | [[File:kahn_fl_image.jpg|250 px|right]] |
| - | |-
| + | Fluorescent proteins have become ubiquitous tools for studying cellular processes, and are frequently used to visualize the dynamics of synthetic networks in cells. To be particularly effective for these applications, fluorescent proteins must feature fast maturation and degradation rates, and these rates must be well-characterized and documented. The 2011 VT iGEM team has worked to find and characterize fluorescent proteins and degradation tags that more quickly degrade them. Here, we present chemical and mathematical models based on two parameters, maturation and degradation rates, which will hopefully contribute to more quantitative characterization and usage of fluorescent reporters in the future, and will inform future efforts in parts characterization. In conducting our experimentation, we tested different fluorescent proteins with degradation tags in <i>Escherichia coli </i> and <i>Saccharomyces cerevisiae</i> using automated fluorescent microscopy techniques, and then worked to determine a comprehensive, accurate mathematical basis for fluorescent protein characterization. |
| - | |
| + | |
| - | ''Tell us more about your project. Give us background. Use this as the abstract of your project. Be descriptive but concise (1-2 paragraphs)''
| + | |
| - | |[[Image:Virginia_Tech_team.png|right|frame|Your team picture]]
| + | |
| - | |-
| + | |
| - | |
| + | |
| - | |align="center"|[[Team:Virginia_Tech | Team Example]]
| + | |
| - | |}
| + | |
| | | | |
| - | <!--- The Mission, Experiments --->
| |
| | | | |
| - | {| style="color:#1b2c8a;background-color:#0c6;" cellpadding="3" cellspacing="1" border="1" bordercolor="#fff" width="62%" align="center"
| + | |
| - | !align="center"|[[Team:Virginia_Tech|Home]]
| + | Our team consists of 10 undergraduates from Virginia Tech, UNC Chapel Hill, James Madison University, The University of Maryland, Bluefield State College, and The College of William & Mary. We would like to thank Dr. Jean Peccoud and the Synthetic Biology Group at the Virginia Bioinformatics Institute for lab space, financial support, and guidance. |
| - | !align="center"|[[Team:Virginia_Tech/Team|Team]]
| + | |
| - | !align="center"|[https://igem.org/Team.cgi?year=2011&team_name=Virginia_Tech Official Team Profile]
| + | |
| - | !align="center"|[[Team:Virginia_Tech/Project|Project]]
| + | |
| - | !align="center"|[[Team:Virginia_Tech/Parts|Parts Submitted to the Registry]]
| + | |
| - | !align="center"|[[Team:Virginia_Tech/Modeling|Modeling]]
| + | |
| - | !align="center"|[[Team:Virginia_Tech/Notebook|Notebook]]
| + | |
| - | !align="center"|[[Team:Virginia_Tech/Safety|Safety]]
| + | |
| - | !align="center"|[[Team:Virginia_Tech/Attributions|Attributions]]
| + | |
| - | |}
| + | |
Characterization of Fluorescent Reporters:
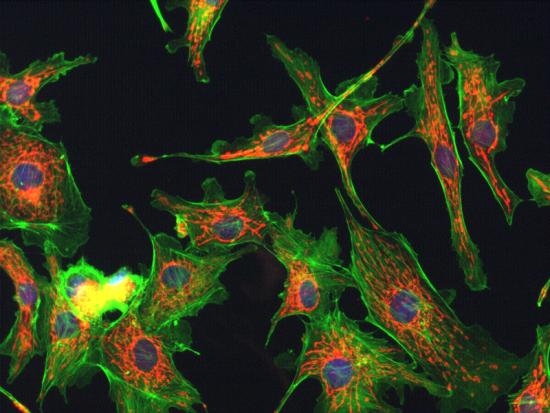
Fluorescent proteins have become ubiquitous tools for studying cellular processes, and are frequently used to visualize the dynamics of synthetic networks in cells. To be particularly effective for these applications, fluorescent proteins must feature fast maturation and degradation rates, and these rates must be well-characterized and documented. The 2011 VT iGEM team has worked to find and characterize fluorescent proteins and degradation tags that more quickly degrade them. Here, we present chemical and mathematical models based on two parameters, maturation and degradation rates, which will hopefully contribute to more quantitative characterization and usage of fluorescent reporters in the future, and will inform future efforts in parts characterization. In conducting our experimentation, we tested different fluorescent proteins with degradation tags in Escherichia coli and Saccharomyces cerevisiae using automated fluorescent microscopy techniques, and then worked to determine a comprehensive, accurate mathematical basis for fluorescent protein characterization.
Our team consists of 10 undergraduates from Virginia Tech, UNC Chapel Hill, James Madison University, The University of Maryland, Bluefield State College, and The College of William & Mary. We would like to thank Dr. Jean Peccoud and the Synthetic Biology Group at the Virginia Bioinformatics Institute for lab space, financial support, and guidance.
|
 "
"