Team:BU Wellesley Software/Notebook/ShannonNotebook
From 2011.igem.org
(Difference between revisions)
| (34 intermediate revisions not shown) | |||
| Line 1: | Line 1: | ||
| + | <html> | ||
| + | <head> | ||
| + | <title>BU-Wellesley iGEM Team: Welcome</title> | ||
| + | <meta http-equiv="Content-Type" content="text/html; charset=utf-8"> | ||
| + | <script src="http://cdn.jquerytools.org/1.2.5/full/jquery.tools.min.js?foo"></script> | ||
| + | <style type="text/css"> | ||
| + | /*hide default igem banner and reformat style into blank slate*/ | ||
| + | #globalWrapper {width: 100%;} | ||
| + | #top-section {width: 100%; height:30px; border:none;} | ||
| + | #p-logo {display:none;} | ||
| + | #search-controls {display:none;} | ||
| + | #menubar a {color:#000000;} | ||
| + | #menubar a:hover{text-decoration:none; color:#52749C;} | ||
| + | .left-menu {background-color:#FFFFFF; margin:5px 0px 0px 0px; padding:0;} | ||
| + | .left-menu ul {background-color:#FFFFFF; margin:0; padding:0;} | ||
| + | .right-menu ul li a {background-color:#FFFFFF;} | ||
| + | .printfooter {display:none;} | ||
| + | #footer-box {border:none;} | ||
| + | #catlinks {display:none;} | ||
| + | .firstHeading {display:none;} | ||
| + | #content {width: 100%; border:none;} | ||
| + | #bodyContent {border:none;} | ||
| + | |||
| + | /*actual content styles*/ | ||
| + | body {width: 800px; margin:auto;} | ||
| + | |||
| + | #bu-wellesley_wiki_content {height:auto; line-height:100%;} | ||
| + | /*#bu-wellesley_wiki_content a {color:#69d01d;}*/ | ||
| + | #bu-wellesley_wiki_content a:hover {text-decoration:none; color:#3d3f3c;} | ||
| + | |||
| + | .navbar li {color: #ffffff;} | ||
| + | .navbar li a {color: #ffffff;} | ||
| + | .navbar li a:hover {background:#69d01d; color: #ffffff;} | ||
| + | |||
| + | /*only use for current page content header (i.e. Team, G-nomeSurferPro, etc)*/ | ||
| + | H6 { | ||
| + | font-family: Helvetica; | ||
| + | text-transform: uppercase; | ||
| + | text-decoration: none; | ||
| + | text-align: left; | ||
| + | color: #3d3f3c; | ||
| + | font-size: 32pt; | ||
| + | } | ||
| + | |||
| + | </style> | ||
| + | |||
| + | <link rel="stylesheet" type="text/css" href="http://cs.wellesley.edu/~hcilab/iGEM_wiki/css/Team.css"> | ||
| + | |||
| + | </head> | ||
| + | <body class="basiclayout"> | ||
| + | <div id="bu-wellesley_wiki_content"> | ||
| + | |||
| + | <p style="text-align:center;"><a href="https://2011.igem.org/Team:BU_Wellesley_Software"><img src="http://cs.wellesley.edu/~hcilab/iGEM_wiki/images/banner.png" width="800px"></a></p> | ||
| + | |||
| + | <ul id="nav"> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Team">Team</a></li> | ||
| + | <li><a href="#">Project</a> | ||
| + | <ul> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Project_Overview">Overview</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Clotho">Clotho</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/G-nomeSurferPro">G-nome Surfer Pro</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/OptimusPrimer">Optimus Primer</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Trumpet">Trumpet</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Puppetshow">Puppetshow</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/eLabNotebook">eLabNotebook</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Wet_Lab">Wet Lab</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Downloads_and_Tutorials">Downloads and Tutorials</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="#">Process</a> | ||
| + | <ul> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Methodology">Methodology</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Safety">Safety</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Notebook">Notebook</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Outreach">Outreach</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Tips">Tips and Tricks</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Gold">Medal Fulfillment</a></li> | ||
| + | <li><a href="#">Additional Info</a> | ||
| + | <ul> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Acknowledgement">Acknowledgement</a></li> | ||
| + | <li><a href="https://2011.igem.org/Team:BU_Wellesley_Software/Social">Fun</a></li> | ||
| + | </ul> | ||
| + | </li> | ||
| + | </ul> | ||
| + | |||
| + | <br> | ||
| + | <h6>Shannon's Notebook</h6> | ||
| + | </body> | ||
| + | </html> | ||
| + | |||
| + | |||
__TOC__ | __TOC__ | ||
| - | {| style="width:800px;background:# | + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#000000;margin-top:5px;" cellspacing="18" |
|style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h5>WEEK 1: 6/06-6/10/2011</h5> | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h5>WEEK 1: 6/06-6/10/2011</h5> | ||
|- | |- | ||
| Line 16: | Line 109: | ||
|} | |} | ||
| - | {| style="width:800px;background:# | + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#000000;margin-top:5px;" cellspacing="18" |
|style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h5>WEEK 2: 6/13-6/17/2011</h5> | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h5>WEEK 2: 6/13-6/17/2011</h5> | ||
|- | |- | ||
| Line 31: | Line 124: | ||
|} | |} | ||
| - | {| style="width:800px;background:# | + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#000000;margin-top:5px;" cellspacing="18" |
|style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h5>WEEK 3: 6/20-6/24/2011</h5> | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h5>WEEK 3: 6/20-6/24/2011</h5> | ||
|- | |- | ||
| Line 149: | Line 242: | ||
|} | |} | ||
| - | {| style="width:800px;background:# | + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#000000;margin-top:5px;" cellspacing="18" |
|style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h5>WEEK 4: 6/27-7/01/2011</h5> | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h5>WEEK 4: 6/27-7/01/2011</h5> | ||
|- | |- | ||
| Line 269: | Line 362: | ||
* The 2.5A, B, and C all refer to various samples that were ligated with 2.5 uL of ligase. For example, sample 1A was ligated with 1 uL of ligase. | * The 2.5A, B, and C all refer to various samples that were ligated with 2.5 uL of ligase. For example, sample 1A was ligated with 1 uL of ligase. | ||
| - | * Glycerol stocks were also made from the | + | * Glycerol stocks were also made from the week's successful overnight cultures. |
| + | * Overnight cultures were made using the week's successful transformations. | ||
| + | * To increase DNA concentration in ligations, I tried ethanol precipitation with GFPc. | ||
| + | |} | ||
| + | |||
| + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#000000;margin-top:5px;" cellspacing="18" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h5>WEEK 5: 7/04-7/08/2011</h5> | ||
| + | |- | ||
| + | | | ||
| + | |||
| + | * Transformations were done of various parts in the beginning of the week (including BFP + Term and YFPc + R2000). | ||
| + | * Ligation was also performed on ECFP + Term and EYFP + Term. Transformations were then done using those ligations. | ||
| + | * However, transformations were not successful (no colonies grew). | ||
| + | * Overnight cultures were made. Nanodrop was then done. Yielded decent values (ranging from 25.6-61.9 ng/uL). | ||
| + | |} | ||
| + | |||
| + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#000000;margin-top:5px;" cellspacing="18" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h5>WEEK 6: 7/11-7/15/2011</h5> | ||
| + | |- | ||
| + | | | ||
| + | |||
| + | [[File:sh 7 -12 labeled.png|350px|right|]] | ||
| + | |||
| + | |||
| + | * Restriction digest was done on ECFP and EYFP (both cut with E + S). | ||
| + | * Gel was run later that day of RD products. However, no bands were visible. | ||
| + | * Re-did restriction digest of ECFP and EYFP. Ran a gel. | ||
| + | ** Well 2 (in 12 well comb) contained cut ECFP. | ||
| + | ** Well 3 (in 12 well comb) contained uncut ECFP. | ||
| + | ** Wells 10 and 11 (in 20 well comb) contained cut EYFP. | ||
| + | ** Well 12 (in 20 well comb) contained uncut EYFP. | ||
| + | * Gel extraction was performed later that day. ECFP had a concentration on 41.8 ng/uL and EYFP had a concentration of 18.8 ng/uL. | ||
| + | * To ensure that ligations were not producing false positives, CIP was used on the backbone. | ||
| + | * Ligations were done using a CIP backbone and a non-CIP backbone. | ||
| + | * Transformations were then done using both CIP and non-CIP backbone and sat on the bench top over the weekend. | ||
| + | |} | ||
| + | |||
| + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#000000;margin-top:5px;" cellspacing="18" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h5>WEEK 7: 7/18-7/22/2011</h5> | ||
| + | |- | ||
| + | | | ||
| + | |||
| + | * Transformations from the previous week worked (both CIP and non-CIP). | ||
| + | * Made overnight cultures using the CIP and non-CIP transformations. | ||
| + | * Decided to troubleshoot ligations by trying different ratios and different lengths of time. | ||
| + | * Transformed ligations that were used to troubleshoot. | ||
| + | * Overnight cultures did not grow. Streaked plates from glycerol stocks and made overnight cultures. | ||
| + | * Attempted to ligate ECFP + Term and EYFP + Term. Also transformed the ligations. Put in the 37 C overnight. | ||
| + | |} | ||
| + | |||
| + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#000000;margin-top:5px;" cellspacing="18" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h5>WEEK 8: 7/25-7/29/2011</h5> | ||
| + | |- | ||
| + | | | ||
| + | |||
| + | [[File:07-28-2011 labeled 2.png|350px|right|]] | ||
| + | |||
| + | * Transformations from the previous week seemed to work. Made overnight cultures. Some samples looked clear (some did not appear to have much growth). Yielded poor DNA concentrations (ranging from 2.0 to 19.4). | ||
| + | * Re-ligated ECFP + Term and EYFP + Term. Transformed the samples and put in the 37 C overnight. Ultimately, these transformations did not grow. | ||
| + | * Restriction digest was done on EYFP, ECFP, and Terminator. Gel was run. | ||
| + | ** Well 1: Ladder | ||
| + | ** Well 2: Cut ECFP | ||
| + | ** Well 3: Uncut ECFP | ||
| + | ** Well 4: Cut EYFP | ||
| + | ** Well 5: Uncut EYFP | ||
| + | ** Well 6: Cut Terminator | ||
| + | ** Well 7: Uncut Terminator | ||
| + | * Only band that cut was Terminator. Did gel extraction and Nanodrop. | ||
| + | * Overnight cultues were made from streaked plates. Minipreps were done and results were decent (ranging from 26.7 ng/uL to 42.1 ng/uL). | ||
| + | |} | ||
| + | |||
| + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#000000;margin-top:5px;" cellspacing="18" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h5>WEEK 9: 8/01-8/05/2011</h5> | ||
| + | |- | ||
| + | | | ||
| + | |||
| + | * Restriction digest was done on ECFP, EYFP, and Terminator. Stored in the -20 C overnight. Overnight cultures were made for ECFP and EYFP. | ||
| + | * Minipreps were done for the overnight cultures that were done the previous day. Nanodrop was then done (produced decent values, ranging from 28.3 ng/uL to 33.0 ng/uL | ||
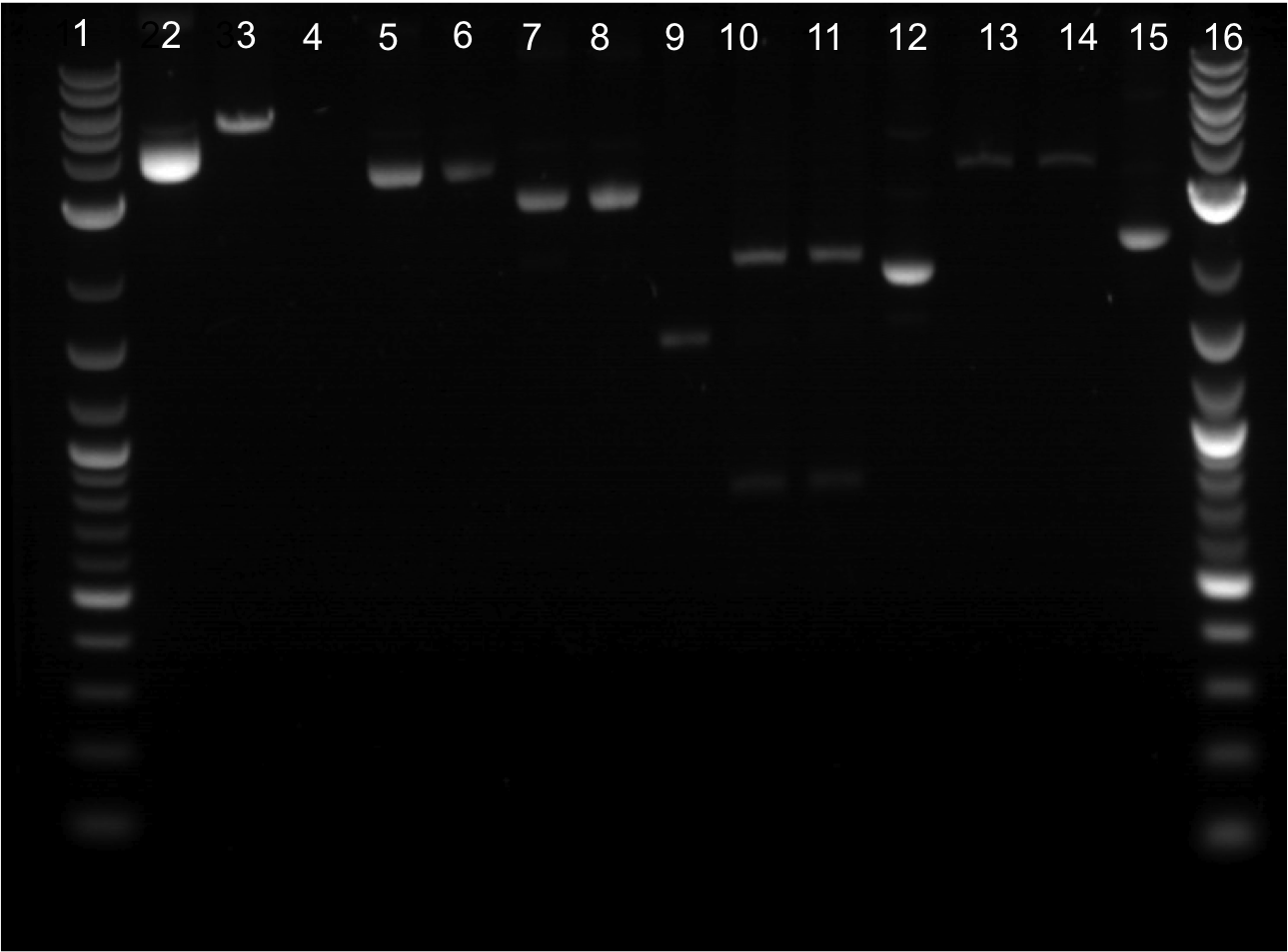
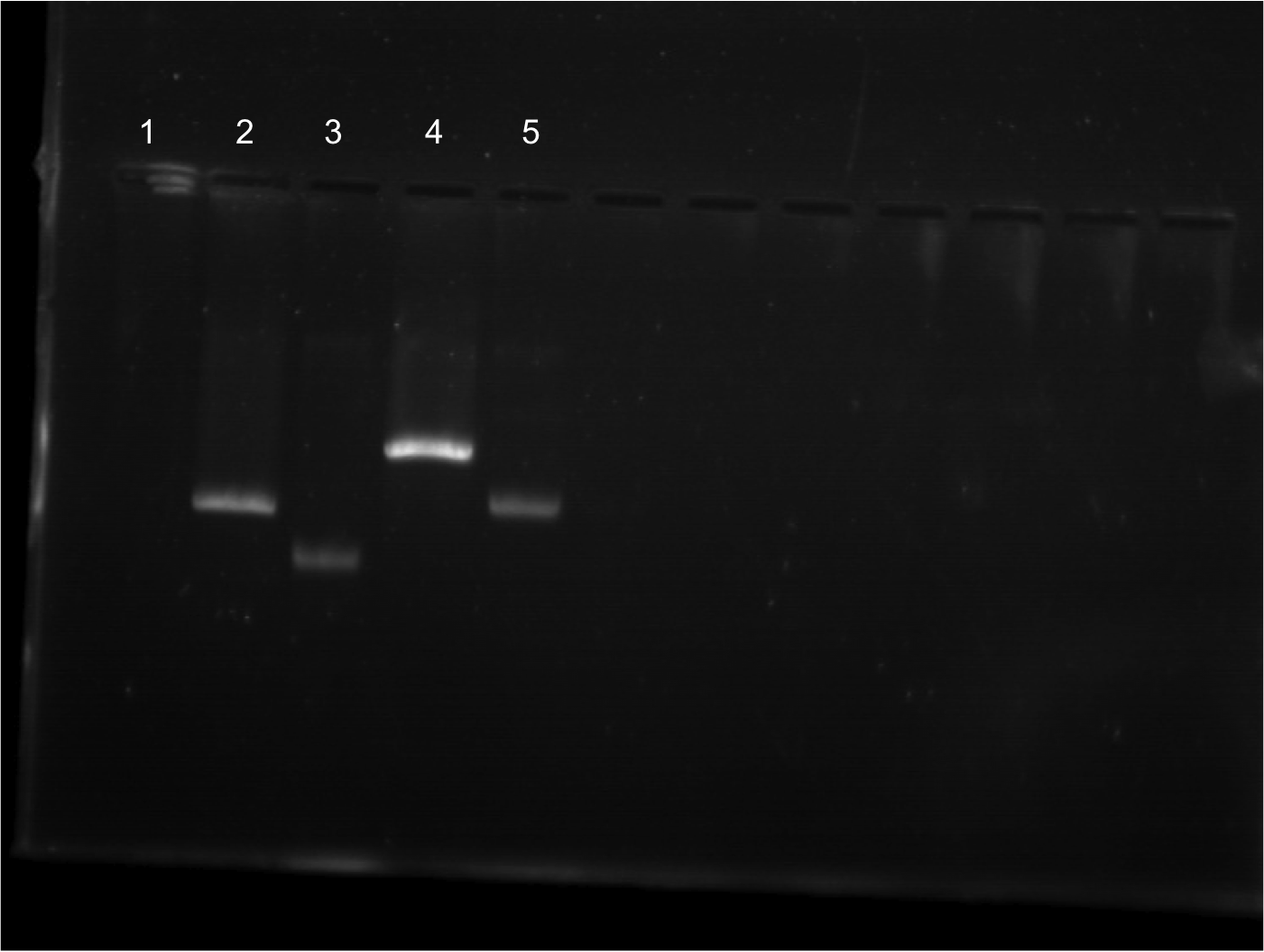
| + | * Ran a gel from 8/01/2011’s restriction digest. Was done in a 20 well comb. | ||
| + | |||
| + | |||
| + | [[File:FINAL FINAL 8.2.png|350px|right|]] | ||
| + | |||
| + | |||
| + | * Lineup for gel | ||
| + | ** Lane 10: Cut EYFP | ||
| + | ** Lane 11: Cut EYFP | ||
| + | ** Lane 12: Uncut EYFP | ||
| + | ** Lane 13: Cut Terminator | ||
| + | ** Lane 14: Cut Terminator | ||
| + | ** Lane 15: Uncut Terminator | ||
| + | |||
| + | * EYFP appeared to cut. Cut out two lower bands and did gel extraction. | ||
| + | * Restriction digest of ECFP and EYFP was performed. This time, I tried using a higher concentration of DNA in the restriction digest. Instead of the usual 500 ng, I tried doubling the concentration to 1000 ng (one sample had 700 ng instead of 1000 ng). Previously, all samples were cut with E + S to ligate with terminator. However, I had not had any success, so I cut with X + P to ligate with RBS instead. This digest was also done overnight. | ||
| + | * Transformations were also done with ECFP (E0020) and EYFP (E0030) from the iGEM plates. Plated 100 uL of dilute cells and then 100 uL of concentrated cells. The next day the plates looked good. Put them in the 4 C. | ||
| + | |} | ||
| + | |||
| + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#000000;margin-top:5px;" cellspacing="18" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h5>WEEK 10: 8/08-8/12/2011</h5> | ||
| + | |- | ||
| + | | | ||
| + | |||
| + | * Overnight cultures were made for 8/05/2011’s transformations. | ||
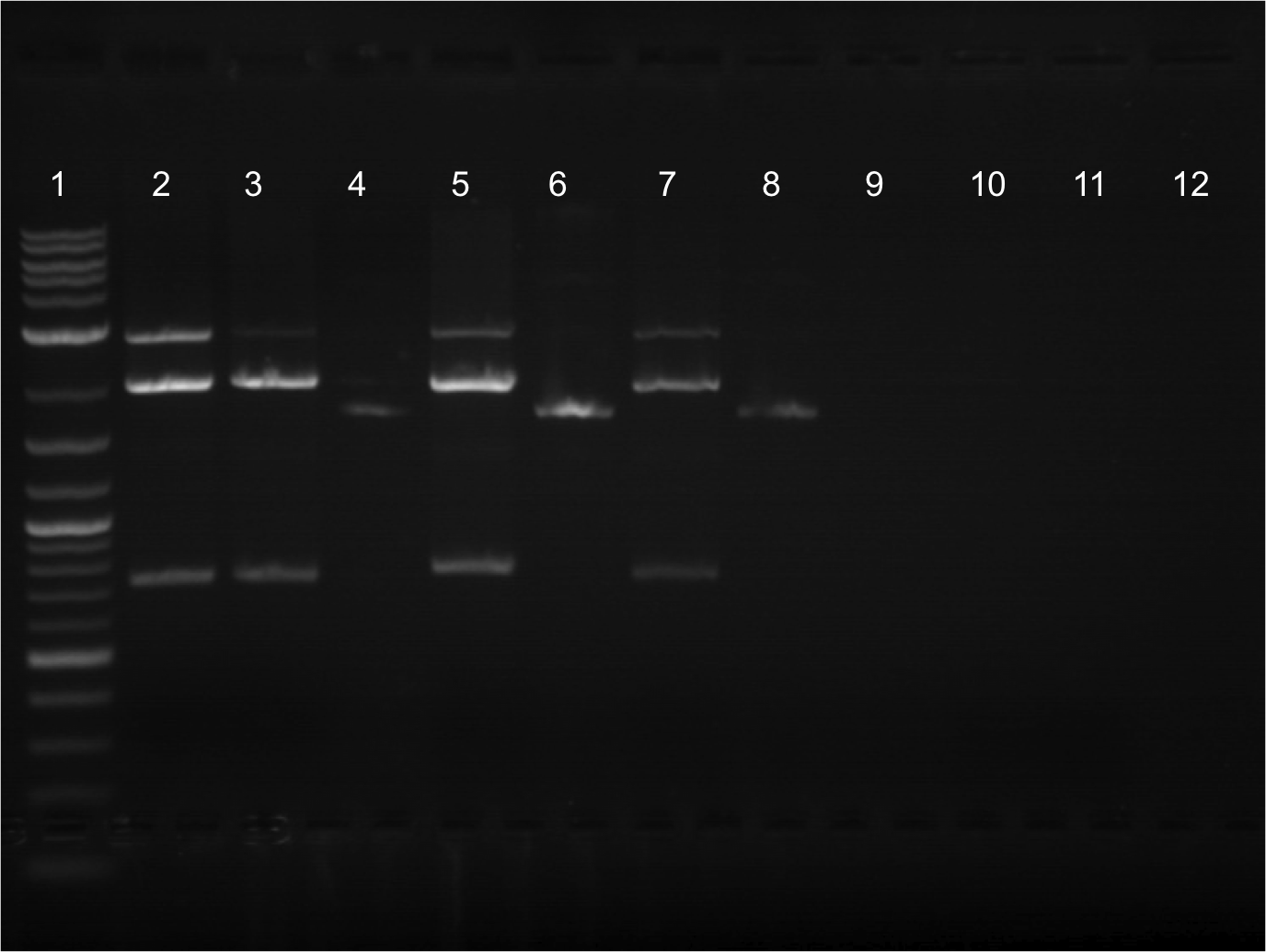
| + | * Gel was run from the restriction digest that was done in the later part of the previous week. All samples cut. Did gel extraction. | ||
| + | |||
| + | [[File:8.8.11 FINAL EDITED.png|350px|right|]] | ||
| + | |||
| + | * Lineup for gel. | ||
| + | ** Lane 1: Ladder | ||
| + | ** Lane 2: Cut E0020.2 | ||
| + | ** Lane 3: Cut E0020 | ||
| + | ** Lane 4: Uncut E0020 | ||
| + | ** Lane 5: Cut E0032.2 | ||
| + | ** Lane 6: Uncut E0032.2 | ||
| + | ** Lane 7: Cut E0032 | ||
| + | ** Lane 8: Uncut | ||
| + | ** Lane 9: Vanessa | ||
| + | ** Lane 10: Vanessa | ||
| + | ** Lane 11: Vanessa | ||
| + | ** Lane 12: empty | ||
| + | |||
| + | |||
| + | * Minipreps were done for overnight cultures that were prepared the day before. Nanodrop results are shown below. | ||
| + | |||
| + | |||
| + | {| class="wikitable" | ||
| + | |- | ||
| + | ! Tube Label | ||
| + | ! Sample | ||
| + | ! ng/uL | ||
| + | |||
| + | |- | ||
| + | | ECFP A1 | ||
| + | | E0020 | ||
| + | | 32.2 | ||
| + | |- | ||
| + | | ECFP A2 | ||
| + | | E0020 | ||
| + | | 28.5 | ||
| + | |- | ||
| + | | ECFP B1 | ||
| + | | E0020 | ||
| + | | 33.8 | ||
| + | |- | ||
| + | | ECFP B2 | ||
| + | | E0020 | ||
| + | | 35.7 | ||
| + | |- | ||
| + | | EYFP A1 | ||
| + | | E0030 | ||
| + | | 71.1 | ||
| + | |- | ||
| + | | EYFP A2 | ||
| + | | E0030 | ||
| + | | 61.0 | ||
| + | |- | ||
| + | | EYFP B1 | ||
| + | | E0030 | ||
| + | | 86.9 | ||
| + | |- | ||
| + | | EYFP B2 | ||
| + | | E0030 | ||
| + | | 47.8 | ||
| + | |} | ||
| + | |||
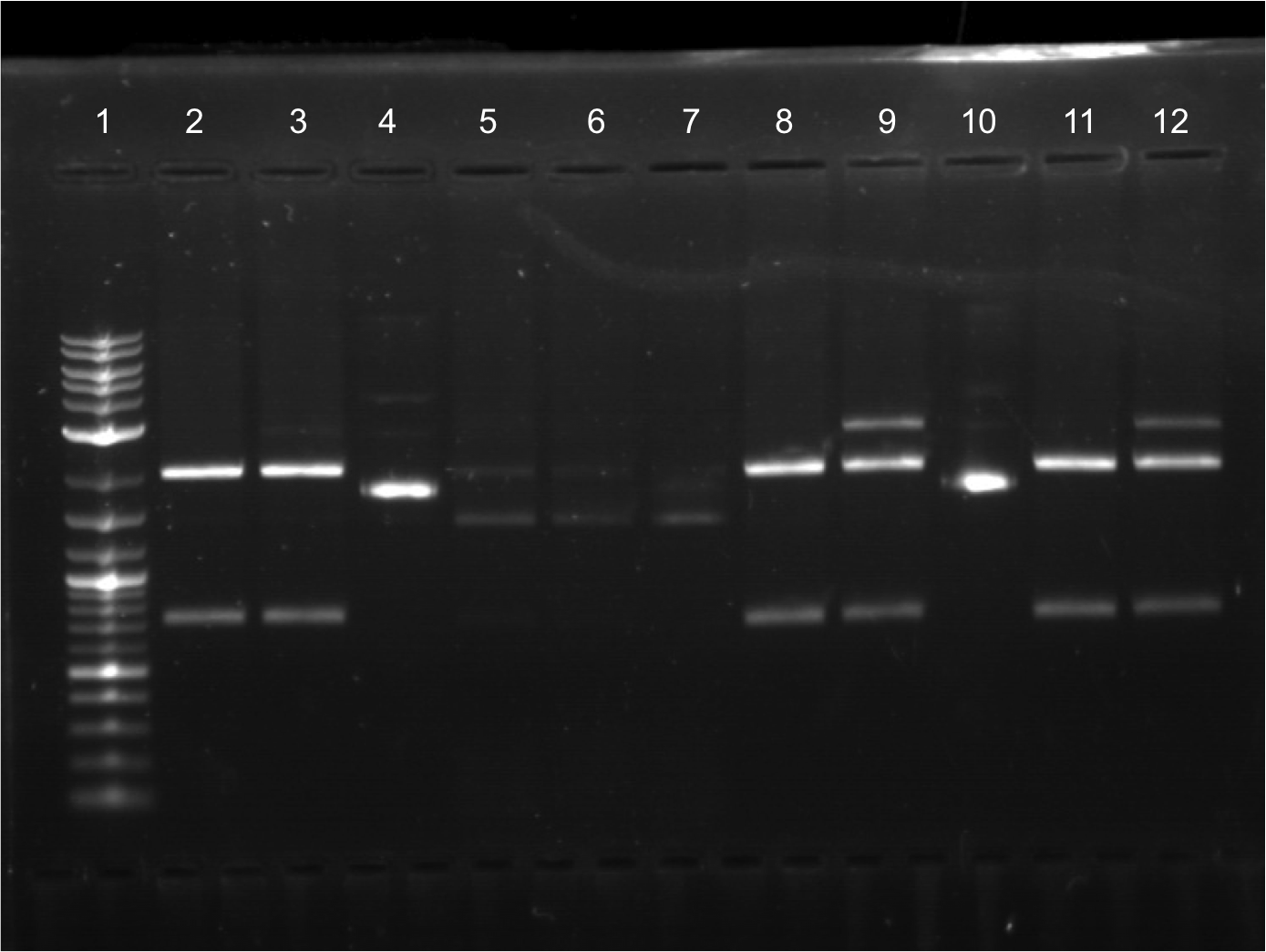
| + | * Did restriction digest on all samples (ECFP A1 – EYFP B2). Did two restriction digest samples per one miniprep sample (one sample cut with second cut with X + P). | ||
| + | * Ran a gel with restriction digest samples. | ||
| + | |||
| + | |||
| + | [[File:8.9.11 FINAL EDITED.png|350px|right|]] | ||
| + | |||
| + | * Lineup for wells: | ||
| + | * 12 well. | ||
| + | ** Lane 1: Ladder | ||
| + | ** Lane 2: Cut ECFP A1, E + S | ||
| + | ** Lane 3: Cut ECFP A1, X + P | ||
| + | ** Lane 4: Uncut ECFP A1 | ||
| + | ** Lane 5: Cut ECFP A2, E + S | ||
| + | ** Lane 6: Cut ECFP A2, X + P | ||
| + | ** Lane 7: Uncut ECFP A2 | ||
| + | ** Lane 8: Cut ECFP B1, E + S | ||
| + | ** Lane 9: Cut ECFP B1, X + P | ||
| + | ** Lane 10: Uncut ECFP B1 | ||
| + | ** Lane 11: Cut ECFP B2, E + S | ||
| + | ** Lane 12: Cut ECFP B2, X + P | ||
| + | |||
| + | |||
| + | [[File:8.9 gel.png|350px|right|]] | ||
| + | |||
| + | * 20 well. | ||
| + | ** Lane 1: Ladder | ||
| + | ** Lane 2: Uncut ECFP B2 | ||
| + | ** Lane 3: Cut EYFP A1, E + S | ||
| + | ** Lane 4: Cut EYFP A1, X + P | ||
| + | ** Lane 5: Uncut EYFP A1 | ||
| + | ** Lane 6: Cut EYFP A2, E + S | ||
| + | ** Lane 7: Cut EYFP A2, X + P | ||
| + | ** Lane 8: Uncut EYFP A2 | ||
| + | ** Lane 9: Cut EYFP B1, E + S | ||
| + | ** Lane 10: Cut EYFP B1, X + P | ||
| + | ** Lane 11: Uncut EYFP B1 | ||
| + | ** Lane 12: Cut EYFP B2, E + S | ||
| + | ** Lane 13: Cut EYFP B2, X + P | ||
| + | ** Lane 14: Uncut EYFP B2 | ||
| + | ** Lane 15: empty | ||
| + | ** Lane 16: empty | ||
| + | ** Lane 17: empty | ||
| + | ** Lane 18: empty | ||
| + | ** Lane 19: empty | ||
| + | ** Lane 20: Ladder | ||
| + | * Continued with gel extraction, Nanodrop, ligation, and transformation. | ||
| + | |} | ||
| + | |||
| + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#000000;margin-top:5px;" cellspacing="18" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h5>WEEK 11: 8/15-8/19/2011</h5> | ||
| + | |- | ||
| + | | | ||
| + | |||
| + | * All ligations/transformations worked except for ECFP + B0015 and EYFP + B0015. Overnight cultures were made for all successful transformations. | ||
| + | * Minipreps were done and Nanodrop followed. Nanodrop yielded decent results (values ranging from 21.2-53.4 ng/uL). | ||
| + | * 08/17-08/21: out of Boston. | ||
| + | |} | ||
| + | |||
| + | {| style="width:800px;background:#F0FFF0;text-align:justify;font-family: helvetica, arial, sans-serif;color:#000000;margin-top:5px;" cellspacing="18" | ||
| + | |style="font-family: helvetica, arial, sans-serif;font-size:1em;color:#ea8828;"|<h5>WEEK 12: 8/22-8/26/2011</h5> | ||
| + | |- | ||
| + | | | ||
| + | |||
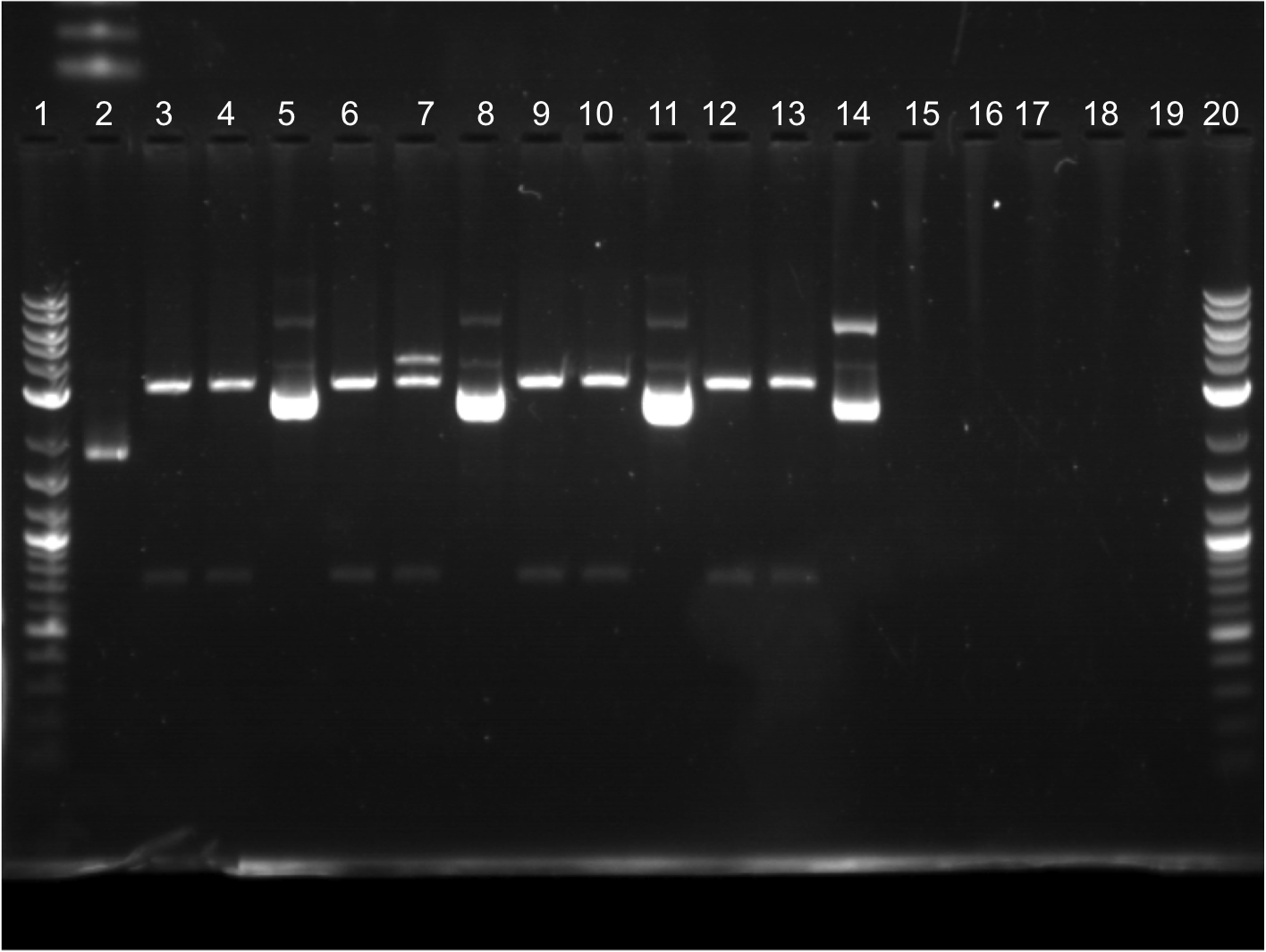
| + | * Restriction digest was performed on all ECFP and EYFP plus RBS combinations. Terminator and B0034 were also cut. | ||
| + | * All samples were run on a gel. All bands appeared to be cut. | ||
| + | |||
| + | |||
| + | |||
| + | [[File:8.23 first 12 well.png|350px|right|]] | ||
| + | |||
| + | * 1st 12 well: | ||
| + | ** Lane 1: Ladder | ||
| + | ** Lane 2: Cut ECFP + J61100C | ||
| + | ** Lane 3: Uncut ECFP + J61100C | ||
| + | ** Lane 4: Cut ECFP + J61127A | ||
| + | ** Lane 5: Uncut ECFP + J61127A | ||
| + | ** Lane 6: Cut EYFP + J61100C | ||
| + | ** Lane 7: Uncut EYFP + J61100C | ||
| + | ** Lane 8: Cut EYFP + J61127C | ||
| + | ** Lane 9: Uncut EYFP + J61127C | ||
| + | ** Lane 10: empty | ||
| + | ** Lane 11: empty | ||
| + | ** Lane 12: Ladder | ||
| + | |||
| + | |||
| + | [[File:8.23 second 12 well.png|350px|right|]] | ||
| + | |||
| + | * 2nd 12 well: | ||
| + | ** Lane 1: Ladder | ||
| + | ** Lane 2: Cut B0034 | ||
| + | ** Lane 3: Uncut B0034 | ||
| + | ** Lane 4: Cut B0015 | ||
| + | ** Lane 5: Uncut B0015 | ||
| + | ** Lane 6: empty | ||
| + | ** Lane 7: empty | ||
| + | ** Lane 8: empty | ||
| + | ** Lane 9: empty | ||
| + | ** Lane 10: empty | ||
| + | ** Lane 11: empty | ||
| + | ** Lane 12: empty | ||
| + | * Gel extraction and Nanodrop followed. | ||
| + | * All samples were then ligated with terminator and transformed. Previously cut EYFP and ECFP were ligated with B0034 and transformed. | ||
| + | |||
| + | |||
| + | [[File:IMG_0273.jpg|350px|left|]] [[File:IMG_0274.jpg|350px|right|]] | ||
| + | [[File:IMG_0275.jpg|350px|left|]] [[File:IMG_0278.jpg|350px|right|]] | ||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |||
| + | |} | ||
Latest revision as of 01:27, 29 September 2011
Shannon's Notebook
WEEK 1: 6/06-6/10/2011 |
|
WEEK 2: 6/13-6/17/2011 |
|
WEEK 3: 6/20-6/24/2011 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
WEEK 4: 6/27-7/01/2011 | ||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||||
|
WEEK 5: 7/04-7/08/2011 |
|
WEEK 6: 7/11-7/15/2011 |
|
|
WEEK 7: 7/18-7/22/2011 |
|
WEEK 8: 7/25-7/29/2011 |
|
WEEK 9: 8/01-8/05/2011 |
|
WEEK 10: 8/08-8/12/2011 | |||||||||||||||||||||||||||
|
WEEK 11: 8/15-8/19/2011 |
|
WEEK 12: 8/22-8/26/2011 |
|
 "
"