Team:Berkeley/Parts
From 2011.igem.org
Gabriellopez (Talk | contribs) |
Gabriellopez (Talk | contribs) |
||
| (66 intermediate revisions not shown) | |||
| Line 43: | Line 43: | ||
</div> | </div> | ||
| - | <div class="row" | + | <div class="row"> |
| - | + | <div class="col12" id="spacer3"></div> | |
| - | + | <div class="row-end"> </div> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
| + | <div class="row"> | ||
| + | <div class="col12"> <img src="https://static.igem.org/mediawiki/2011/0/09/SummaryPartsHeader.jpg" width="980"> </div> </div> | ||
| + | <div class="row-end" ></div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | <div class="row"> | ||
| + | <div class="col12-3"> | ||
| - | + | <b># of parts submitted:</b> | |
| - | + | <ul> | |
| - | + | <li>35 Regulatory promoters</li> | |
| - | + | <li>2 Ribosome Binding Sites</li> | |
| - | + | <li>2 Composite expression modules</li> | |
| + | <li>2 Coding proteins</li> | ||
| + | </ul> | ||
| + | <b>Total:</b> 42 Parts <br> <br> | ||
| + | <b>Assembly Method:</b> BglBricks <br><br> | ||
| + | <b>Accomplishments:</b> <br> | ||
| + | <ul> | ||
| + | <li>All 42 parts have been submitted to the registry and can be found <a href="http://partsregistry.org/cgi/partsdb/pgroup.cgi?pgroup=iGEM2011&group=Berkeley">HERE</a></li> | ||
| + | <li>We submitted two new well-characterized BioBrick Parts in vector pSB1AC3 according to RFC10</li> | ||
| + | <li>Compiled a set of 35 stress promoters that future iGEMers can utilize as regulatory elements</li> | ||
| + | </ul> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| + | <br> | ||
</div> | </div> | ||
| - | + | <div class="row-end" ></div> | |
| - | + | ||
| - | <div class="row | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
</div> | </div> | ||
| - | |||
| - | |||
<div class="row"> | <div class="row"> | ||
| Line 128: | Line 84: | ||
</div> | </div> | ||
| - | </div> | + | <div class="row"> |
| - | </ | + | <div class="col12"> <img src="https://static.igem.org/mediawiki/2011/f/fb/PartssubHeader2.jpg" width="980"> </div> </div> |
| + | <div class="row-end" ></div> | ||
| + | <div class="row"> | ||
| + | <div class="col12-3"> | ||
| + | <h3>Function</h3> | ||
| + | Stress promoters respond differently to various types of stress that is placed on the cell. We wanted a promoter that would be downregulated in the presence of membrane stress caused by the overexpression of ToxR fusion proteins. The design principle was to express ToxR fusion proteins under this promoter such that the stress it caused would downregulate its production. | ||
| + | <h3>Assembly</h3> | ||
| + | The 34 stress promoters, identified from microarray data (Moen, 2009), were PCRed from E. coli MG1655 Genomic DNA. Basic parts were made in plasmids with pUC origins. A constitutive promoter (Pcon) was also cloned into a basic part as a control for characterization. | ||
| + | <h3>Characterization</h3> | ||
| + | <p>The pool of 35 promoters was assembled in front of a rbs.sfGFP reporter gene, and the construct was transformed into E. coli MC1061. The transformed cells were subjected to generalized stresses (heat, cold, acid, base, salt) overnight, and their resulting fluorescence was measured.</p> | ||
| + | <center> | ||
| + | <b><img src="https://static.igem.org/mediawiki/2011/0/03/Stress_Conditions.jpg" width= "400" height="250" align="center"></b></center> | ||
| + | <b></b> | ||
| + | <h3>Data</h3> | ||
| + | <p>Downregulation in response to stress, resulting in decreased fluorescence, was desired. Tecan plate measurements showed initial downregulation in some of the stress promoeters, with the cold condition yielding the best results. Further flow cytometry experiments confirmed the stress-responsive downregulation shown by the Tecan. In the figures below, we show fluorescence relative to that of the constitutive promoter in both conditions. Here we show our cold condition (25 degrees C) vs our control condition (37 degrees C). We find that that these chosen 4 promoters have downregulated activity from the environmental stress we put on the cells.</p> | ||
| + | <img src="https://static.igem.org/mediawiki/2011/f/fd/Cytometrydata.png" height="275" ><img src="https://static.igem.org/mediawiki/2011/3/3d/Tecandata.png" height="285" align="right"> | ||
| + | </div> | ||
| + | <div class="row-end" ></div> | ||
| + | </div> | ||
| + | <div class="row"> | ||
| + | <div class="col12" id="spacer3"></div> | ||
| + | <div class="row-end"> </div> | ||
| + | </div> | ||
| + | <div class="row"> | ||
| + | <div class="col12"> <img src="https://static.igem.org/mediawiki/2011/b/b6/PartssubHeader3.jpg" width="980"> </div> </div> | ||
| + | <div class="row-end" ></div> | ||
| + | <div class="row"> | ||
| + | <div class="col12-3"> | ||
| + | <h3>Function</h3> | ||
| + | The expression plasmid consists of a stress promoter regulating the production of ToxR. This construct allows us to express enough ToxR to allow for the functionality we desire and at the same time have minimal affect on bacterial growth rate. | ||
| + | <h3>Assembly</h3> | ||
| + | The ToxR was PCR amplified out of V. cholerae genomic DNA, digested, and ligated in front of the leucine zipper IILK. A library of potential stress promoters and an RBS library were put in front of the ToxR fusion protein and the library was screened for healthy, transcriptionally active clones. We found a combination of the rffGH stress promoter and an rbs variant gave us the functionality we desired. This construct was then used as our template for our other ToxR fusion proteins. Note: for our ToxR chimeras, we switched from an IILK constitutive dimerizing periplasmic domain to Lambda-Rep and MukF constitutive dimerizing periplasmic domains after we identified the stress promoter because they provided higher levels fluorescence in our experiments. For this reason, we collected data using ToxR fusions of Lambda-Rep and MukF. | ||
| - | + | <br><br><center><img src="https://static.igem.org/mediawiki/2011/1/14/ToxRLamRep.png" height="120" align="center"></center> | |
| - | + | ||
| - | + | ||
| - | < | + | |
| - | < | + | <h3>Characterization</h3> |
| + | For characterization, we made fusion proteins of ToxR with Lamda Repressor and MukF proteins. Both of these proteins dimerize in wild-type and we expect them to help ToxR dimerize as well. We transformed ToxR-LambdaRep driven by Pstress into our reporter strain (see below for more details). In parallel, we also transformed our original constructs of ToxR-lambdaRep driven by Pbad into our reporter strains. Each construct was grown in 100 mg/ml, 50 mg/ml, 25 mg/ml, 12 mg/ml, 6 mg/ml 3 mg/ml, 1.5 mg/ml, and 0 mg/ml arabinose. For our control, a known nontoxic plasmid was transformed into the cells that should not activate our reporter strain. | ||
| - | < | + | <h3>Data</h3> |
| - | + | As shown in the graph of Pbad-LambdaRep and Pbad-MukF, cells did not grow up well in the varying levels of arabinose. The relative fluorescence graphs also supported the conclusion that our constructs were toxic to the cells. | |
| - | + | ||
| - | < | + | <br><br/> |
| + | Our solution to the high toxicity of our ToxR chimeras was to have a promoter that would down regulate transcription when the cell was under stress. As shown in the Pstress graphs of LambdaRep and MukF, cells grew to a much higher OD and fluorescence especially in Pstress-LambdaRep compared to the control. All levels of arabinose showed similar results. | ||
| - | < | + | <center> |
| - | < | + | <img src="https://static.igem.org/mediawiki/2011/7/73/Pbad_lamdarep.png" height="350"> <br> |
| - | + | <img src="https://static.igem.org/mediawiki/2011/d/d9/Pbad_mukF.png" height="350"> <br> | |
| - | + | <img src="https://static.igem.org/mediawiki/2011/d/d4/Pstress_lamdarep.png" height="350"> <br> | |
| - | + | <img src="https://static.igem.org/mediawiki/2011/2/26/Pstress_mukF.png" height="350"> <br> | |
| - | + | <img src="https://static.igem.org/mediawiki/2011/e/e5/Control.png" height="350"> | |
| - | + | </center> | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | + | ||
| - | </ | + | |
| - | |||
| - | |||
| - | |||
</div> | </div> | ||
| - | |||
| - | |||
| - | |||
<div class="row-end" ></div> | <div class="row-end" ></div> | ||
</div> | </div> | ||
| - | |||
<div class="row"> | <div class="row"> | ||
| Line 194: | Line 161: | ||
</div> | </div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
<div class="row"> | <div class="row"> | ||
| - | <div class="col12 | + | <div class="col12"> <img src="https://static.igem.org/mediawiki/2011/d/dc/PartssubHeader4.jpg" width="980"> </div> </div> |
| + | <div class="row-end" ></div> | ||
| - | |||
| - | |||
<div class="row"> | <div class="row"> | ||
| - | <div class="col12 | + | <div class="col12-3"> |
| - | + | ||
| - | + | ||
| + | <h3>Function</h3> | ||
| + | The reporter construct give a fluorescent output that correlates to the amount of transcriptional activity ToxR promotes on Pctx (it's wild type promoter). Since transcriptional activity by ToxR dimers depends on the strength of dimerization, the level of fluorescence can be correlated to how tightly the ToxR dimerizes. | ||
| - | + | <h3>Assembly</h3> | |
| - | + | ToxR’s native target, the ctx promoter (Pctx), was PCRed out of Vibrio cholerae genomic DNA and made into a basic part. The promoter was then placed upstream of an rbs.sfGFP part using 2AB assembly. | |
| - | + | ||
| - | < | + | <br><br><center><img src="https://static.igem.org/mediawiki/2011/2/27/PctxGFP.png" height="100" align="center"></center> |
| - | |||
| - | < | + | <h3>Characterization</h3> |
| + | By cotransforming this reporter plasmid and the expression plasmid (see above) we were able to measure the strength of ctx promoter (Pctx) induction by ToxR. | ||
| + | <h3>Data</h3> | ||
| + | See Expression Plasmid Data above. | ||
| - | + | </div> | |
| - | <div class="row-end"></div> | + | <div class="row-end" ></div> |
| - | + | </div> | |
| - | + | ||
<div class="row"> | <div class="row"> | ||
| - | <div class="col12" id=" | + | <div class="col12" id="spacer3"></div> |
<div class="row-end"> </div> | <div class="row-end"> </div> | ||
</div> | </div> | ||
| - | + | <div class="row"> | |
| - | <div class=" | + | <div class="col12"> <img src="https://static.igem.org/mediawiki/2011/0/04/PartssubHeader5.jpg" width="980"> </div> </div> |
| - | + | <div class="row-end" ></div> | |
| - | <div class=" | + | |
| - | <div class="row-end"></div> | + | |
| - | + | ||
| - | + | ||
<div class="row"> | <div class="row"> | ||
| - | <div class=" | + | <div class="col12-3"> |
| + | <h3>Function</h3> | ||
| + | The human estrogen receptor is known to dimerize around estradiol. By fusing the human estrogen receptor to ToxR, we can create an estradiol biosensor and potentially an estradiol-inducible system in E. coli. | ||
| + | <h3>Assembly</h3> | ||
| + | The estrogen receptor was refactored to optimally express in E. coli and assembled using designed oligos and PCA (polymerase cycling assembly). The final protein was made into a basic part. Various sub-sections of the estrogen receptor were PCR amplified as basic parts in order to create truncations that might display different responses to estradiol (see figure). The estrogen receptor was fused to ToxR’s C-terminus with Gly-Ser linkers and thus exposed to the periplasm when expressed in the cell. | ||
| - | < | + | <br><br> <center><img src="https://static.igem.org/mediawiki/2011/2/29/EstrogenToxR.png" height="100"></center> |
| - | < | + | |
| - | < | + | |
| - | + | ||
| + | <h3>Characterization</h3> | ||
| + | In order to characterize the system, the different truncations of estrogen receptor fused to ToxR were tested with a plate reader. In our control condition, no estrogen was added. For the experimental condition, 1uM of estradiol was added to the solution. | ||
| + | <br><br> | ||
| + | <center><img src="https://static.igem.org/mediawiki/2011/f/ff/EstrogenFigureFinal.png" height="370"></center> | ||
| - | + | <h3>Data</h3> | |
| + | Below are the results of the plate reader data for our control and experimental conditions described above. We find that there is no truncation of estrogen receptor that gives an estradiol-inducible system; however, we are optimistic about trying other truncations and ToxR fusions that might lead us to an estrogen biosensor. | ||
| - | < | + | <br><br> <center><img src="https://static.igem.org/mediawiki/2011/9/97/Estrogendata.png" width="800"></center> |
| - | < | + | |
| - | + | ||
| - | + | ||
| - | + | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | + | </div> | |
| - | + | <div class="row-end" ></div> | |
| - | + | ||
| - | + | ||
</div> | </div> | ||
<div class="row"> | <div class="row"> | ||
| - | <div class="col12" id=" | + | <div class="col12" id="spacer3"></div> |
<div class="row-end"> </div> | <div class="row-end"> </div> | ||
</div> | </div> | ||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
| - | |||
</div> | </div> | ||
</html> | </html> | ||
Latest revision as of 00:14, 29 September 2011



- 35 Regulatory promoters
- 2 Ribosome Binding Sites
- 2 Composite expression modules
- 2 Coding proteins
Assembly Method: BglBricks
Accomplishments:
- All 42 parts have been submitted to the registry and can be found HERE
- We submitted two new well-characterized BioBrick Parts in vector pSB1AC3 according to RFC10
- Compiled a set of 35 stress promoters that future iGEMers can utilize as regulatory elements

Function
Stress promoters respond differently to various types of stress that is placed on the cell. We wanted a promoter that would be downregulated in the presence of membrane stress caused by the overexpression of ToxR fusion proteins. The design principle was to express ToxR fusion proteins under this promoter such that the stress it caused would downregulate its production.Assembly
The 34 stress promoters, identified from microarray data (Moen, 2009), were PCRed from E. coli MG1655 Genomic DNA. Basic parts were made in plasmids with pUC origins. A constitutive promoter (Pcon) was also cloned into a basic part as a control for characterization.Characterization
The pool of 35 promoters was assembled in front of a rbs.sfGFP reporter gene, and the construct was transformed into E. coli MC1061. The transformed cells were subjected to generalized stresses (heat, cold, acid, base, salt) overnight, and their resulting fluorescence was measured.

Data
Downregulation in response to stress, resulting in decreased fluorescence, was desired. Tecan plate measurements showed initial downregulation in some of the stress promoeters, with the cold condition yielding the best results. Further flow cytometry experiments confirmed the stress-responsive downregulation shown by the Tecan. In the figures below, we show fluorescence relative to that of the constitutive promoter in both conditions. Here we show our cold condition (25 degrees C) vs our control condition (37 degrees C). We find that that these chosen 4 promoters have downregulated activity from the environmental stress we put on the cells.



Function
The expression plasmid consists of a stress promoter regulating the production of ToxR. This construct allows us to express enough ToxR to allow for the functionality we desire and at the same time have minimal affect on bacterial growth rate.Assembly
The ToxR was PCR amplified out of V. cholerae genomic DNA, digested, and ligated in front of the leucine zipper IILK. A library of potential stress promoters and an RBS library were put in front of the ToxR fusion protein and the library was screened for healthy, transcriptionally active clones. We found a combination of the rffGH stress promoter and an rbs variant gave us the functionality we desired. This construct was then used as our template for our other ToxR fusion proteins. Note: for our ToxR chimeras, we switched from an IILK constitutive dimerizing periplasmic domain to Lambda-Rep and MukF constitutive dimerizing periplasmic domains after we identified the stress promoter because they provided higher levels fluorescence in our experiments. For this reason, we collected data using ToxR fusions of Lambda-Rep and MukF.
Characterization
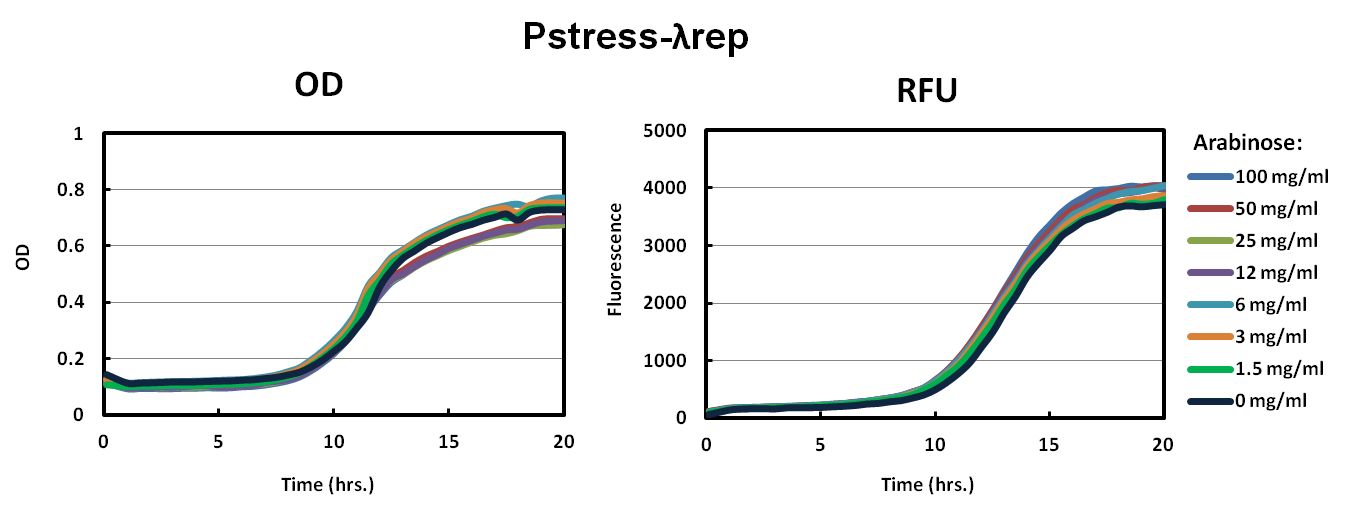
For characterization, we made fusion proteins of ToxR with Lamda Repressor and MukF proteins. Both of these proteins dimerize in wild-type and we expect them to help ToxR dimerize as well. We transformed ToxR-LambdaRep driven by Pstress into our reporter strain (see below for more details). In parallel, we also transformed our original constructs of ToxR-lambdaRep driven by Pbad into our reporter strains. Each construct was grown in 100 mg/ml, 50 mg/ml, 25 mg/ml, 12 mg/ml, 6 mg/ml 3 mg/ml, 1.5 mg/ml, and 0 mg/ml arabinose. For our control, a known nontoxic plasmid was transformed into the cells that should not activate our reporter strain.Data
As shown in the graph of Pbad-LambdaRep and Pbad-MukF, cells did not grow up well in the varying levels of arabinose. The relative fluorescence graphs also supported the conclusion that our constructs were toxic to the cells.Our solution to the high toxicity of our ToxR chimeras was to have a promoter that would down regulate transcription when the cell was under stress. As shown in the Pstress graphs of LambdaRep and MukF, cells grew to a much higher OD and fluorescence especially in Pstress-LambdaRep compared to the control. All levels of arabinose showed similar results.





Function
The reporter construct give a fluorescent output that correlates to the amount of transcriptional activity ToxR promotes on Pctx (it's wild type promoter). Since transcriptional activity by ToxR dimers depends on the strength of dimerization, the level of fluorescence can be correlated to how tightly the ToxR dimerizes.Assembly
ToxR’s native target, the ctx promoter (Pctx), was PCRed out of Vibrio cholerae genomic DNA and made into a basic part. The promoter was then placed upstream of an rbs.sfGFP part using 2AB assembly.
Characterization
By cotransforming this reporter plasmid and the expression plasmid (see above) we were able to measure the strength of ctx promoter (Pctx) induction by ToxR.Data
See Expression Plasmid Data above.
Function
The human estrogen receptor is known to dimerize around estradiol. By fusing the human estrogen receptor to ToxR, we can create an estradiol biosensor and potentially an estradiol-inducible system in E. coli.Assembly
The estrogen receptor was refactored to optimally express in E. coli and assembled using designed oligos and PCA (polymerase cycling assembly). The final protein was made into a basic part. Various sub-sections of the estrogen receptor were PCR amplified as basic parts in order to create truncations that might display different responses to estradiol (see figure). The estrogen receptor was fused to ToxR’s C-terminus with Gly-Ser linkers and thus exposed to the periplasm when expressed in the cell.
Characterization
In order to characterize the system, the different truncations of estrogen receptor fused to ToxR were tested with a plate reader. In our control condition, no estrogen was added. For the experimental condition, 1uM of estradiol was added to the solution.
Data
Below are the results of the plate reader data for our control and experimental conditions described above. We find that there is no truncation of estrogen receptor that gives an estradiol-inducible system; however, we are optimistic about trying other truncations and ToxR fusions that might lead us to an estrogen biosensor.
 "
"